+ データを開く
データを開く
- 基本情報
基本情報
| 登録情報 | データベース: EMDB / ID: EMD-2990 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| タイトル | Structure of Target Of Rapapmycin Complex 2 (TORC2) from Saccharomyces cerevisiae | |||||||||
 マップデータ マップデータ | Reconstruction of yeast TORC2 - filtered at 26 Angstroms resolution (FSC 0.143 criterion after gold standard refinement). Contour level provided by author - The map was generated from negative stain data, and we determined the correct contour level to display the map to be 2.9 in Pymol. | |||||||||
 試料 試料 |
| |||||||||
 キーワード キーワード | TOR kinase complex / TORC2 / cell growth / rapamycin | |||||||||
| 生物種 |  | |||||||||
| 手法 | 単粒子再構成法 / ネガティブ染色法 / 解像度: 26.0 Å | |||||||||
 データ登録者 データ登録者 | Gaubitz C / Oliveira TM / Prouteau M / Leitner A / Karuppasamy M / Konstantinidou G / Rispal D / Eltschinger S / Robinson GC / Thore S ...Gaubitz C / Oliveira TM / Prouteau M / Leitner A / Karuppasamy M / Konstantinidou G / Rispal D / Eltschinger S / Robinson GC / Thore S / Aebersold R / Schaffitzel C / Loewith R | |||||||||
 引用 引用 |  ジャーナル: Mol Cell / 年: 2015 ジャーナル: Mol Cell / 年: 2015タイトル: Molecular Basis of the Rapamycin Insensitivity of Target Of Rapamycin Complex 2. 著者: Christl Gaubitz / Taiana M Oliveira / Manoel Prouteau / Alexander Leitner / Manikandan Karuppasamy / Georgia Konstantinidou / Delphine Rispal / Sandra Eltschinger / Graham C Robinson / ...著者: Christl Gaubitz / Taiana M Oliveira / Manoel Prouteau / Alexander Leitner / Manikandan Karuppasamy / Georgia Konstantinidou / Delphine Rispal / Sandra Eltschinger / Graham C Robinson / Stéphane Thore / Ruedi Aebersold / Christiane Schaffitzel / Robbie Loewith /    要旨: Target of Rapamycin (TOR) plays central roles in the regulation of eukaryote growth as the hub of two essential multiprotein complexes: TORC1, which is rapamycin-sensitive, and the lesser ...Target of Rapamycin (TOR) plays central roles in the regulation of eukaryote growth as the hub of two essential multiprotein complexes: TORC1, which is rapamycin-sensitive, and the lesser characterized TORC2, which is not. TORC2 is a key regulator of lipid biosynthesis and Akt-mediated survival signaling. In spite of its importance, its structure and the molecular basis of its rapamycin insensitivity are unknown. Using crosslinking-mass spectrometry and electron microscopy, we determined the architecture of TORC2. TORC2 displays a rhomboid shape with pseudo-2-fold symmetry and a prominent central cavity. Our data indicate that the C-terminal part of Avo3, a subunit unique to TORC2, is close to the FKBP12-rapamycin-binding domain of Tor2. Removal of this sequence generated a FKBP12-rapamycin-sensitive TORC2 variant, which provides a powerful tool for deciphering TORC2 function in vivo. Using this variant, we demonstrate a role for TORC2 in G2/M cell-cycle progression. | |||||||||
| 履歴 |
|
- 構造の表示
構造の表示
| ムービー |
 ムービービューア ムービービューア |
|---|---|
| 構造ビューア | EMマップ:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| 添付画像 |
- ダウンロードとリンク
ダウンロードとリンク
-EMDBアーカイブ
| マップデータ |  emd_2990.map.gz emd_2990.map.gz | 3.6 MB |  EMDBマップデータ形式 EMDBマップデータ形式 | |
|---|---|---|---|---|
| ヘッダ (付随情報) |  emd-2990-v30.xml emd-2990-v30.xml emd-2990.xml emd-2990.xml | 10 KB 10 KB | 表示 表示 |  EMDBヘッダ EMDBヘッダ |
| 画像 |  emd_2990.tif emd_2990.tif | 739.6 KB | ||
| アーカイブディレクトリ |  http://ftp.pdbj.org/pub/emdb/structures/EMD-2990 http://ftp.pdbj.org/pub/emdb/structures/EMD-2990 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-2990 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-2990 | HTTPS FTP |
-検証レポート
| 文書・要旨 |  emd_2990_validation.pdf.gz emd_2990_validation.pdf.gz | 216.5 KB | 表示 |  EMDB検証レポート EMDB検証レポート |
|---|---|---|---|---|
| 文書・詳細版 |  emd_2990_full_validation.pdf.gz emd_2990_full_validation.pdf.gz | 215.7 KB | 表示 | |
| XML形式データ |  emd_2990_validation.xml.gz emd_2990_validation.xml.gz | 4.7 KB | 表示 | |
| アーカイブディレクトリ |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-2990 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-2990 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-2990 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-2990 | HTTPS FTP |
-関連構造データ
- リンク
リンク
| EMDBのページ |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- マップ
マップ
| ファイル |  ダウンロード / ファイル: emd_2990.map.gz / 形式: CCP4 / 大きさ: 3.7 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) ダウンロード / ファイル: emd_2990.map.gz / 形式: CCP4 / 大きさ: 3.7 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 注釈 | Reconstruction of yeast TORC2 - filtered at 26 Angstroms resolution (FSC 0.143 criterion after gold standard refinement). Contour level provided by author - The map was generated from negative stain data, and we determined the correct contour level to display the map to be 2.9 in Pymol. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 投影像・断面図 | 画像のコントロール
画像は Spider により作成 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| ボクセルのサイズ | X=Y=Z: 4.5 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
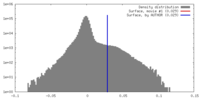
| 密度 |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 対称性 | 空間群: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 詳細 | EMDB XML:
CCP4マップ ヘッダ情報:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-添付データ
- 試料の構成要素
試料の構成要素
-全体 : Yeast Target of Rapamycin Complex 2
| 全体 | 名称: Yeast Target of Rapamycin Complex 2 |
|---|---|
| 要素 |
|
-超分子 #1000: Yeast Target of Rapamycin Complex 2
| 超分子 | 名称: Yeast Target of Rapamycin Complex 2 / タイプ: sample / ID: 1000 / 詳細: TORC2 complex, which consists of six subunits / 集合状態: dimer / Number unique components: 1 |
|---|---|
| 分子量 | 実験値: 1.4 MDa / 理論値: 1.4 MDa / 手法: Size exclusion |
-分子 #1: Yeast TORC2
| 分子 | 名称: Yeast TORC2 / タイプ: protein_or_peptide / ID: 1 詳細: TORC2 is composed of two copies of: Tor2 kinase, Lst8, Avo1, Avo2, Avo3, Bit61/2 コピー数: 1 / 集合状態: 2 / 組換発現: No |
|---|---|
| 由来(天然) | 生物種:  細胞中の位置: Plasma membrane MCT (membrane compartment containing TORC2) domain |
| 分子量 | 実験値: 1.4 MDa / 理論値: 1.4 MDa |
-実験情報
-構造解析
| 手法 | ネガティブ染色法 |
|---|---|
 解析 解析 | 単粒子再構成法 |
| 試料の集合状態 | particle |
- 試料調製
試料調製
| 濃度 | 0.01 mg/mL |
|---|---|
| 緩衝液 | pH: 7.5 詳細: 50mM HEPES pH 7.5, 5 mM CHAPS, 300 mM KCl, 0,5 mM DTT |
| 染色 | タイプ: NEGATIVE 詳細: Grids with adsorbed protein floated on 2% w/v uranyl acetate for 1 min. |
| グリッド | 詳細: 300 mesh grid |
| 凍結 | 凍結剤: NONE / 装置: OTHER |
- 電子顕微鏡法
電子顕微鏡法
| 顕微鏡 | FEI/PHILIPS CM120T |
|---|---|
| 日付 | 2012年4月10日 |
| 撮影 | カテゴリ: CCD フィルム・検出器のモデル: TVIPS TEMCAM-F415 (4k x 4k) 実像数: 200 / 詳細: 200 micrographs (100 tilt pairs) |
| Tilt angle min | 0 |
| 電子線 | 加速電圧: 120 kV / 電子線源:  FIELD EMISSION GUN FIELD EMISSION GUN |
| 電子光学系 | 照射モード: FLOOD BEAM / 撮影モード: BRIGHT FIELD / 倍率(公称値): 26000 |
| 試料ステージ | 試料ホルダーモデル: SIDE ENTRY, EUCENTRIC / Tilt angle max: 45 |
- 画像解析
画像解析
| 詳細 | A total of 8,762 RCT pairs were picked manually with tiltpicker (Voss et al, 2009). The additional untilted particles were picked with e2boxer.py (Ludtke, 2010). The 3D reconstruction was calculated with the Xmipp ML tomo and refined with Xmipp MLF 3D (Scheres et al., 2008) |
|---|---|
| CTF補正 | 詳細: each micrograph |
| 最終 再構成 | 想定した対称性 - 点群: C1 (非対称) / アルゴリズム: OTHER / 解像度のタイプ: BY AUTHOR / 解像度: 26.0 Å / 解像度の算出法: OTHER / ソフトウェア - 名称: Xmipp, Relion / 使用した粒子像数: 24979 |
| 最終 2次元分類 | クラス数: 500 |
 ムービー
ムービー コントローラー
コントローラー













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)