+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Hepatitis B virus capsid bound to importin alpha1 | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Capsid formed by Cp183 when bound to importin alpha1 / VIRUS LIKE PARTICLE | |||||||||
| Function / homology |  Function and homology information Function and homology informationmicrotubule-dependent intracellular transport of viral material towards nucleus / T=4 icosahedral viral capsid / viral penetration into host nucleus / host cell / host cell cytoplasm / symbiont entry into host cell / structural molecule activity / DNA binding / RNA binding / extracellular region Similarity search - Function | |||||||||
| Biological species |   Hepatitis B virus Hepatitis B virus | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.8 Å | |||||||||
 Authors Authors | Yang R / Cingolani G | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Sci Adv / Year: 2024 Journal: Sci Adv / Year: 2024Title: Structural basis for nuclear import of hepatitis B virus (HBV) nucleocapsid core. Authors: Ruoyu Yang / Ying-Hui Ko / Fenglin Li / Ravi K Lokareddy / Chun-Feng David Hou / Christine Kim / Shelby Klein / Santiago Antolínez / Juan F Marín / Carolina Pérez-Segura / Martin F ...Authors: Ruoyu Yang / Ying-Hui Ko / Fenglin Li / Ravi K Lokareddy / Chun-Feng David Hou / Christine Kim / Shelby Klein / Santiago Antolínez / Juan F Marín / Carolina Pérez-Segura / Martin F Jarrold / Adam Zlotnick / Jodi A Hadden-Perilla / Gino Cingolani /  Abstract: Nuclear import of the hepatitis B virus (HBV) nucleocapsid is essential for replication that occurs in the nucleus. The ~360-angstrom HBV capsid translocates to the nuclear pore complex (NPC) as an ...Nuclear import of the hepatitis B virus (HBV) nucleocapsid is essential for replication that occurs in the nucleus. The ~360-angstrom HBV capsid translocates to the nuclear pore complex (NPC) as an intact particle, hijacking human importins in a reaction stimulated by host kinases. This paper describes the mechanisms of HBV capsid recognition by importins. We found that importin α1 binds a nuclear localization signal (NLS) at the far end of the HBV coat protein Cp183 carboxyl-terminal domain (CTD). This NLS is exposed to the capsid surface through a pore at the icosahedral quasi-sixfold vertex. Phosphorylation at serine-155, serine-162, and serine-170 promotes CTD compaction but does not affect the affinity for importin α1. The binding of 30 importin α1/β1 augments HBV capsid diameter to ~620 angstroms, close to the maximum size trafficable through the NPC. We propose that phosphorylation favors CTD externalization and prompts its compaction at the capsid surface, exposing the NLS to importins. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_29858.map.gz emd_29858.map.gz | 394.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-29858-v30.xml emd-29858-v30.xml emd-29858.xml emd-29858.xml | 13.7 KB 13.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_29858_fsc.xml emd_29858_fsc.xml | 17 KB | Display |  FSC data file FSC data file |
| Images |  emd_29858.png emd_29858.png | 145.6 KB | ||
| Filedesc metadata |  emd-29858.cif.gz emd-29858.cif.gz | 5.2 KB | ||
| Others |  emd_29858_half_map_1.map.gz emd_29858_half_map_1.map.gz emd_29858_half_map_2.map.gz emd_29858_half_map_2.map.gz | 385.5 MB 388.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-29858 http://ftp.pdbj.org/pub/emdb/structures/EMD-29858 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-29858 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-29858 | HTTPS FTP |
-Validation report
| Summary document |  emd_29858_validation.pdf.gz emd_29858_validation.pdf.gz | 1.1 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_29858_full_validation.pdf.gz emd_29858_full_validation.pdf.gz | 1.1 MB | Display | |
| Data in XML |  emd_29858_validation.xml.gz emd_29858_validation.xml.gz | 25.1 KB | Display | |
| Data in CIF |  emd_29858_validation.cif.gz emd_29858_validation.cif.gz | 33.2 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-29858 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-29858 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-29858 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-29858 | HTTPS FTP |
-Related structure data
| Related structure data |  8g8yMC  7umiC  8g5vC  8g6vC  8gcnC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_29858.map.gz / Format: CCP4 / Size: 421.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_29858.map.gz / Format: CCP4 / Size: 421.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.108 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_29858_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
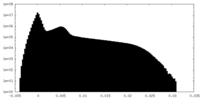
| Projections & Slices |
| ||||||||||||
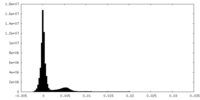
| Density Histograms |
-Half map: #2
| File | emd_29858_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
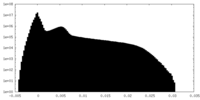
| Projections & Slices |
| ||||||||||||
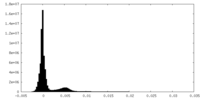
| Density Histograms |
- Sample components
Sample components
-Entire : Capsid formed by Cp183 when importin alpha1 is incorporated
| Entire | Name: Capsid formed by Cp183 when importin alpha1 is incorporated |
|---|---|
| Components |
|
-Supramolecule #1: Capsid formed by Cp183 when importin alpha1 is incorporated
| Supramolecule | Name: Capsid formed by Cp183 when importin alpha1 is incorporated type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:   Hepatitis B virus Hepatitis B virus |
-Macromolecule #1: Core protein Cp183
| Macromolecule | Name: Core protein Cp183 / type: protein_or_peptide / ID: 1 / Number of copies: 12 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Hepatitis B virus Hepatitis B virus |
| Molecular weight | Theoretical: 16.328719 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MDIDPYKEFG ATVELLSFLP SDFFPSVRDL LDTASALYRE ALESPEHCSP HHTALRQAIL CWGELMTLAT WVGVNLEDPA SRDLVVSYV NTNMGLKFRQ LLWFHISCLT FGRETVIEYL VSFGVWIRTP PAYRPPNAPI LSTLP UniProtKB: Capsid protein |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.25 µm / Nominal defocus min: 0.75 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)