[English] 日本語
 Yorodumi
Yorodumi- EMDB-29357: Human IMPDH2 mutant - L245P, treated with ATP, IMP, and NAD+; fil... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Human IMPDH2 mutant - L245P, treated with ATP, IMP, and NAD+; filament assembly interface reconstruction | |||||||||
 Map data Map data | Map from density modification. Used to build model. | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Filament / Dehydrogenase / CBS domain / Bateman domain / purine biosynthesis / OXIDOREDUCTASE | |||||||||
| Function / homology |  Function and homology information Function and homology information'de novo' XMP biosynthetic process / lymphocyte proliferation / Purine ribonucleoside monophosphate biosynthesis / IMP dehydrogenase activity / IMP dehydrogenase / GMP biosynthetic process / Azathioprine ADME / GTP biosynthetic process / peroxisomal membrane / cellular response to interleukin-4 ...'de novo' XMP biosynthetic process / lymphocyte proliferation / Purine ribonucleoside monophosphate biosynthesis / IMP dehydrogenase activity / IMP dehydrogenase / GMP biosynthetic process / Azathioprine ADME / GTP biosynthetic process / peroxisomal membrane / cellular response to interleukin-4 / circadian rhythm / secretory granule lumen / ficolin-1-rich granule lumen / Potential therapeutics for SARS / nucleotide binding / Neutrophil degranulation / DNA binding / RNA binding / extracellular exosome / extracellular region / membrane / nucleus / metal ion binding / cytosol / cytoplasm Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.0 Å | |||||||||
 Authors Authors | O'Neill AG / Kollman JM | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Elife / Year: 2018 Journal: Elife / Year: 2018Title: New tools for automated high-resolution cryo-EM structure determination in RELION-3. Authors: Jasenko Zivanov / Takanori Nakane / Björn O Forsberg / Dari Kimanius / Wim Jh Hagen / Erik Lindahl / Sjors Hw Scheres /    Abstract: Here, we describe the third major release of RELION. CPU-based vector acceleration has been added in addition to GPU support, which provides flexibility in use of resources and avoids memory ...Here, we describe the third major release of RELION. CPU-based vector acceleration has been added in addition to GPU support, which provides flexibility in use of resources and avoids memory limitations. Reference-free autopicking with Laplacian-of-Gaussian filtering and execution of jobs from python allows non-interactive processing during acquisition, including 2D-classification, model generation and 3D-classification. Per-particle refinement of CTF parameters and correction of estimated beam tilt provides higher resolution reconstructions when particles are at different heights in the ice, and/or coma-free alignment has not been optimal. Ewald sphere curvature correction improves resolution for large particles. We illustrate these developments with publicly available data sets: together with a Bayesian approach to beam-induced motion correction it leads to resolution improvements of 0.2-0.7 Å compared to previous RELION versions. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_29357.map.gz emd_29357.map.gz | 12.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-29357-v30.xml emd-29357-v30.xml emd-29357.xml emd-29357.xml | 21.9 KB 21.9 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_29357.png emd_29357.png | 103.6 KB | ||
| Masks |  emd_29357_msk_1.map emd_29357_msk_1.map | 244.1 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-29357.cif.gz emd-29357.cif.gz | 6.6 KB | ||
| Others |  emd_29357_additional_1.map.gz emd_29357_additional_1.map.gz emd_29357_half_map_1.map.gz emd_29357_half_map_1.map.gz emd_29357_half_map_2.map.gz emd_29357_half_map_2.map.gz | 226.6 MB 188 MB 188 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-29357 http://ftp.pdbj.org/pub/emdb/structures/EMD-29357 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-29357 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-29357 | HTTPS FTP |
-Validation report
| Summary document |  emd_29357_validation.pdf.gz emd_29357_validation.pdf.gz | 761.8 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_29357_full_validation.pdf.gz emd_29357_full_validation.pdf.gz | 761.4 KB | Display | |
| Data in XML |  emd_29357_validation.xml.gz emd_29357_validation.xml.gz | 16 KB | Display | |
| Data in CIF |  emd_29357_validation.cif.gz emd_29357_validation.cif.gz | 18.9 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-29357 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-29357 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-29357 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-29357 | HTTPS FTP |
-Related structure data
| Related structure data |  8fozMC  8fuzC  8g8fC  8g9bC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_29357.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_29357.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Map from density modification. Used to build model. | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.843 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_29357_msk_1.map emd_29357_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
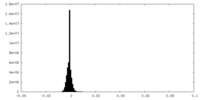
| Density Histograms |
-Additional map: Map from Relion postprocessing.
| File | emd_29357_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Map from Relion postprocessing. | ||||||||||||
| Projections & Slices |
| ||||||||||||
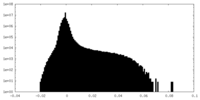
| Density Histograms |
-Half map: #2
| File | emd_29357_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
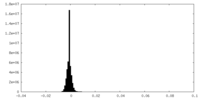
| Density Histograms |
-Half map: #1
| File | emd_29357_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
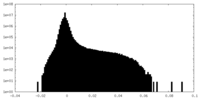
| Density Histograms |
- Sample components
Sample components
-Entire : Filament of Inosine-5'-monophosphate dehydrogenase 2 - L245P muta...
| Entire | Name: Filament of Inosine-5'-monophosphate dehydrogenase 2 - L245P mutant, bound to ATP, IMP, and NAD+ |
|---|---|
| Components |
|
-Supramolecule #1: Filament of Inosine-5'-monophosphate dehydrogenase 2 - L245P muta...
| Supramolecule | Name: Filament of Inosine-5'-monophosphate dehydrogenase 2 - L245P mutant, bound to ATP, IMP, and NAD+ type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Inosine-5'-monophosphate dehydrogenase 2
| Macromolecule | Name: Inosine-5'-monophosphate dehydrogenase 2 / type: protein_or_peptide / ID: 1 / Number of copies: 8 / Enantiomer: LEVO / EC number: IMP dehydrogenase |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 56.464453 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: SEFELMADYL ISGGTSYVPD DGLTAQQLFN CGDGLTYNDF LILPGYIDFT ADQVDLTSAL TKKITLKTPL VSSPMDTVTE AGMAIAMAL TGGIGFIHHN CTPEFQANEV RKVKKYEQGF ITDPVVLSPK DRVRDVFEAK ARHGFCGIPI TDTGRMGSRL V GIISSRDI ...String: SEFELMADYL ISGGTSYVPD DGLTAQQLFN CGDGLTYNDF LILPGYIDFT ADQVDLTSAL TKKITLKTPL VSSPMDTVTE AGMAIAMAL TGGIGFIHHN CTPEFQANEV RKVKKYEQGF ITDPVVLSPK DRVRDVFEAK ARHGFCGIPI TDTGRMGSRL V GIISSRDI DFLKEEEHDC FLEEIMTKRE DLVVAPAGIT LKEANEILQR SKKGKLPIVN EDDELVAIIA RTDLKKNRDY PL ASKDAKK QLPCGAAIGT HEDDKYRLDL LAQAGVDVVV LDSSQGNSIF QINMIKYIKD KYPNLQVIGG NVVTAAQAKN LID AGVDAL RVGMGSGSIC ITQEVLACGR PQATAVYKVS EYARRFGVPV IADGGIQNVG HIAKALALGA STVMMGSLLA ATTE APGEY FFSDGIRLKK YRGMGSLDAM DKHLSSQNRY FSEADKIKVA QGVSGAVQDK GSIHKFVPYL IAGIQHSCQD IGAKS LTQV RAMMYSGELK FEKRTSSAQV EGGVHSLHSY EKRLF UniProtKB: Inosine-5'-monophosphate dehydrogenase 2 |
-Macromolecule #2: INOSINIC ACID
| Macromolecule | Name: INOSINIC ACID / type: ligand / ID: 2 / Number of copies: 8 / Formula: IMP |
|---|---|
| Molecular weight | Theoretical: 348.206 Da |
| Chemical component information | 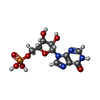 ChemComp-I: |
-Macromolecule #3: NICOTINAMIDE-ADENINE-DINUCLEOTIDE
| Macromolecule | Name: NICOTINAMIDE-ADENINE-DINUCLEOTIDE / type: ligand / ID: 3 / Number of copies: 8 / Formula: NAD |
|---|---|
| Molecular weight | Theoretical: 663.425 Da |
| Chemical component information |  ChemComp-NAD: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | filament |
- Sample preparation
Sample preparation
| Buffer | pH: 7 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Number real images: 2648 / Average exposure time: 3.0 sec. / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 0.27 µm / Nominal defocus min: 1.71 µm / Nominal magnification: 105000 |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT |
|---|---|
| Output model |  PDB-8foz: |
 Movie
Movie Controller
Controller








 X (Sec.)
X (Sec.) Y (Row.)
Y (Row.) Z (Col.)
Z (Col.)