[English] 日本語
 Yorodumi
Yorodumi- EMDB-29848: Human IMPDH2 mutant - L245P, treated with ATP, IMP, and NAD+; ext... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
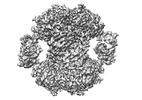
| Title | Human IMPDH2 mutant - L245P, treated with ATP, IMP, and NAD+; extended filament segment reconstruction | |||||||||
 Map data Map data | Map from PHENIX Density Modification. Used to build model. | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Filament / Dehydrogenase / CBS domain / Bateman domain / purine biosynthesis / OXIDOREDUCTASE | |||||||||
| Function / homology |  Function and homology information Function and homology information'de novo' XMP biosynthetic process / Purine ribonucleoside monophosphate biosynthesis / lymphocyte proliferation / IMP dehydrogenase / IMP dehydrogenase activity / GMP biosynthetic process / Azathioprine ADME / peroxisomal membrane / GTP biosynthetic process / cellular response to interleukin-4 ...'de novo' XMP biosynthetic process / Purine ribonucleoside monophosphate biosynthesis / lymphocyte proliferation / IMP dehydrogenase / IMP dehydrogenase activity / GMP biosynthetic process / Azathioprine ADME / peroxisomal membrane / GTP biosynthetic process / cellular response to interleukin-4 / circadian rhythm / secretory granule lumen / ficolin-1-rich granule lumen / Potential therapeutics for SARS / nucleotide binding / Neutrophil degranulation / DNA binding / RNA binding / extracellular exosome / extracellular region / metal ion binding / membrane / nucleus / cytoplasm / cytosol Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.6 Å | |||||||||
 Authors Authors | O'Neill AG / Kollman JM | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation | Journal: J Comput Chem / Year: 2004 Title: UCSF Chimera--a visualization system for exploratory research and analysis. Authors: Eric F Pettersen / Thomas D Goddard / Conrad C Huang / Gregory S Couch / Daniel M Greenblatt / Elaine C Meng / Thomas E Ferrin /  Abstract: The design, implementation, and capabilities of an extensible visualization system, UCSF Chimera, are discussed. Chimera is segmented into a core that provides basic services and visualization, and ...The design, implementation, and capabilities of an extensible visualization system, UCSF Chimera, are discussed. Chimera is segmented into a core that provides basic services and visualization, and extensions that provide most higher level functionality. This architecture ensures that the extension mechanism satisfies the demands of outside developers who wish to incorporate new features. Two unusual extensions are presented: Multiscale, which adds the ability to visualize large-scale molecular assemblies such as viral coats, and Collaboratory, which allows researchers to share a Chimera session interactively despite being at separate locales. Other extensions include Multalign Viewer, for showing multiple sequence alignments and associated structures; ViewDock, for screening docked ligand orientations; Movie, for replaying molecular dynamics trajectories; and Volume Viewer, for display and analysis of volumetric data. A discussion of the usage of Chimera in real-world situations is given, along with anticipated future directions. Chimera includes full user documentation, is free to academic and nonprofit users, and is available for Microsoft Windows, Linux, Apple Mac OS X, SGI IRIX, and HP Tru64 Unix from http://www.cgl.ucsf.edu/chimera/. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_29848.map.gz emd_29848.map.gz | 18.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-29848-v30.xml emd-29848-v30.xml emd-29848.xml emd-29848.xml | 36.1 KB 36.1 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_29848.png emd_29848.png | 104.7 KB | ||
| Masks |  emd_29848_msk_1.map emd_29848_msk_1.map | 244.1 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-29848.cif.gz emd-29848.cif.gz | 8.5 KB | ||
| Others |  emd_29848_additional_1.map.gz emd_29848_additional_1.map.gz emd_29848_half_map_1.map.gz emd_29848_half_map_1.map.gz emd_29848_half_map_2.map.gz emd_29848_half_map_2.map.gz | 224.3 MB 186.9 MB 186.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-29848 http://ftp.pdbj.org/pub/emdb/structures/EMD-29848 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-29848 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-29848 | HTTPS FTP |
-Related structure data
| Related structure data |  8g8fMC  8fozC  8fuzC  8g9bC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_29848.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_29848.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Map from PHENIX Density Modification. Used to build model. | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.843 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_29848_msk_1.map emd_29848_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Map from Relion postprocessing.
| File | emd_29848_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Map from Relion postprocessing. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_29848_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_29848_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Filament of Inosine-5'-monophosphate dehydrogenase 2 - L245P muta...
| Entire | Name: Filament of Inosine-5'-monophosphate dehydrogenase 2 - L245P mutant, bound to ATP, IMP, and NAD+ |
|---|---|
| Components |
|
-Supramolecule #1: Filament of Inosine-5'-monophosphate dehydrogenase 2 - L245P muta...
| Supramolecule | Name: Filament of Inosine-5'-monophosphate dehydrogenase 2 - L245P mutant, bound to ATP, IMP, and NAD+ type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 Details: 5 uM enzyme was mixed with 1 mM ATP, 1 mM MgCl2, 3 mM IMP, and 5 mM NAD+. |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Inosine-5'-monophosphate dehydrogenase 2
| Macromolecule | Name: Inosine-5'-monophosphate dehydrogenase 2 / type: protein_or_peptide / ID: 1 / Number of copies: 8 / Enantiomer: LEVO / EC number: IMP dehydrogenase |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 56.464453 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: SEFELMADYL ISGGTSYVPD DGLTAQQLFN CGDGLTYNDF LILPGYIDFT ADQVDLTSAL TKKITLKTPL VSSPMDTVTE AGMAIAMAL TGGIGFIHHN CTPEFQANEV RKVKKYEQGF ITDPVVLSPK DRVRDVFEAK ARHGFCGIPI TDTGRMGSRL V GIISSRDI ...String: SEFELMADYL ISGGTSYVPD DGLTAQQLFN CGDGLTYNDF LILPGYIDFT ADQVDLTSAL TKKITLKTPL VSSPMDTVTE AGMAIAMAL TGGIGFIHHN CTPEFQANEV RKVKKYEQGF ITDPVVLSPK DRVRDVFEAK ARHGFCGIPI TDTGRMGSRL V GIISSRDI DFLKEEEHDC FLEEIMTKRE DLVVAPAGIT LKEANEILQR SKKGKLPIVN EDDELVAIIA RTDLKKNRDY PL ASKDAKK QLPCGAAIGT HEDDKYRLDL LAQAGVDVVV LDSSQGNSIF QINMIKYIKD KYPNLQVIGG NVVTAAQAKN LID AGVDAL RVGMGSGSIC ITQEVLACGR PQATAVYKVS EYARRFGVPV IADGGIQNVG HIAKALALGA STVMMGSLLA ATTE APGEY FFSDGIRLKK YRGMGSLDAM DKHLSSQNRY FSEADKIKVA QGVSGAVQDK GSIHKFVPYL IAGIQHSCQD IGAKS LTQV RAMMYSGELK FEKRTSSAQV EGGVHSLHSY EKRLF UniProtKB: Inosine-5'-monophosphate dehydrogenase 2 |
-Macromolecule #2: ADENOSINE-5'-TRIPHOSPHATE
| Macromolecule | Name: ADENOSINE-5'-TRIPHOSPHATE / type: ligand / ID: 2 / Number of copies: 16 / Formula: ATP |
|---|---|
| Molecular weight | Theoretical: 507.181 Da |
| Chemical component information |  ChemComp-ATP: |
-Macromolecule #3: INOSINIC ACID
| Macromolecule | Name: INOSINIC ACID / type: ligand / ID: 3 / Number of copies: 8 / Formula: IMP |
|---|---|
| Molecular weight | Theoretical: 348.206 Da |
| Chemical component information | 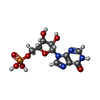 ChemComp-I: |
-Macromolecule #4: NICOTINAMIDE-ADENINE-DINUCLEOTIDE
| Macromolecule | Name: NICOTINAMIDE-ADENINE-DINUCLEOTIDE / type: ligand / ID: 4 / Number of copies: 8 / Formula: NAD |
|---|---|
| Molecular weight | Theoretical: 663.425 Da |
| Chemical component information |  ChemComp-NAD: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | filament |
- Sample preparation
Sample preparation
| Buffer | pH: 7 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Number real images: 2648 / Average exposure time: 3.0 sec. / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 0.27 µm / Nominal defocus min: 1.71 µm / Nominal magnification: 105000 |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT |
|---|---|
| Output model |  PDB-8g8f: |
 Movie
Movie Controller
Controller







 X (Sec.)
X (Sec.) Y (Row.)
Y (Row.) Z (Col.)
Z (Col.)