[English] 日本語
 Yorodumi
Yorodumi- EMDB-29291: Nodavirus RNA replication protein A polymerase domain, local refi... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Nodavirus RNA replication protein A polymerase domain, local refinement | |||||||||
 Map data Map data | Nodavirus RNA replication protein A polymerase domain, local refinement. | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Nodavirus RNA replication protein A polymerase domain / RNA dependent RNA polymerase / VIRAL PROTEIN | |||||||||
| Function / homology | Nodavirus methyltransferase domain / Nodavirus Vmethyltransferase / host cell mitochondrial outer membrane / RNA-directed RNA polymerase / RNA-directed RNA polymerase activity / DNA/RNA polymerase superfamily / membrane / RNA-directed RNA polymerase Function and homology information Function and homology information | |||||||||
| Biological species |  Flock House virus Flock House virus | |||||||||
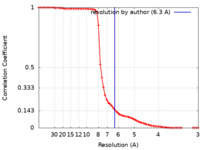
| Method | single particle reconstruction / cryo EM / Resolution: 6.3 Å | |||||||||
 Authors Authors | Zhan H / Unchwaniwala N / Rebolledo Viveros A / Pennington J / Horswill M / Broadberry R / Myers J / den Boon J / Grant T / Ahlquist P | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2023 Journal: Proc Natl Acad Sci U S A / Year: 2023Title: Nodavirus RNA replication crown architecture reveals proto-crown precursor and viral protein A conformational switching. Authors: Hong Zhan / Nuruddin Unchwaniwala / Andrea Rebolledo-Viveros / Janice Pennington / Mark Horswill / Roma Broadberry / Jonathan Myers / Johan A den Boon / Timothy Grant / Paul Ahlquist /  Abstract: Positive-strand RNA viruses replicate their genomes in virus-induced membrane vesicles, and the resulting RNA replication complexes are a major target for virus control. Nodavirus studies first ...Positive-strand RNA viruses replicate their genomes in virus-induced membrane vesicles, and the resulting RNA replication complexes are a major target for virus control. Nodavirus studies first revealed viral RNA replication proteins forming a 12-fold symmetric "crown" at the vesicle opening to the cytosol, an arrangement recently confirmed to extend to distantly related alphaviruses. Using cryoelectron microscopy (cryo-EM), we show that mature nodavirus crowns comprise two stacked 12-mer rings of multidomain viral RNA replication protein A. Each ring contains an ~19 nm circle of C-proximal polymerase domains, differentiated by strikingly diverged positions of N-proximal RNA capping/membrane binding domains. The lower ring is a "proto-crown" precursor that assembles prior to RNA template recruitment, RNA synthesis, and replication vesicle formation. In this proto-crown, the N-proximal segments interact to form a toroidal central floor, whose 3.1 Å resolution structure reveals many mechanistic details of the RNA capping/membrane binding domains. In the upper ring, cryo-EM fitting indicates that the N-proximal domains extend radially outside the polymerases, forming separated, membrane-binding "legs." The polymerase and N-proximal domains are connected by a long linker accommodating the conformational switch between the two rings and possibly also polymerase movements associated with RNA synthesis and nonsymmetric electron density in the lower center of mature crowns. The results reveal remarkable viral protein multifunctionality, conformational flexibility, and evolutionary plasticity and insights into (+)RNA virus replication and control. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_29291.map.gz emd_29291.map.gz | 25.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-29291-v30.xml emd-29291-v30.xml emd-29291.xml emd-29291.xml | 18.2 KB 18.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_29291_fsc.xml emd_29291_fsc.xml | 6.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_29291.png emd_29291.png | 71 KB | ||
| Filedesc metadata |  emd-29291.cif.gz emd-29291.cif.gz | 6.4 KB | ||
| Others |  emd_29291_half_map_1.map.gz emd_29291_half_map_1.map.gz emd_29291_half_map_2.map.gz emd_29291_half_map_2.map.gz | 25 MB 25 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-29291 http://ftp.pdbj.org/pub/emdb/structures/EMD-29291 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-29291 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-29291 | HTTPS FTP |
-Related structure data
| Related structure data |  8fmbMC  8fm9C  8fmaC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_29291.map.gz / Format: CCP4 / Size: 27 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_29291.map.gz / Format: CCP4 / Size: 27 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Nodavirus RNA replication protein A polymerase domain, local refinement. | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.5 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Nodavirus RNA replication protein A polymerase domain, local...
| File | emd_29291_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Nodavirus RNA replication protein A polymerase domain, local refinement, half map1. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Nodavirus RNA replication protein A polymerase domain, local...
| File | emd_29291_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Nodavirus RNA replication protein A polymerase domain, local refinement, half map2. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Nodavirus RNA replication protein A polymerase domain, local refi...
| Entire | Name: Nodavirus RNA replication protein A polymerase domain, local refinement |
|---|---|
| Components |
|
-Supramolecule #1: Nodavirus RNA replication protein A polymerase domain, local refi...
| Supramolecule | Name: Nodavirus RNA replication protein A polymerase domain, local refinement type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Flock House virus Flock House virus |
-Macromolecule #1: RNA-directed RNA polymerase
| Macromolecule | Name: RNA-directed RNA polymerase / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO / EC number: RNA-directed RNA polymerase |
|---|---|
| Source (natural) | Organism:  Flock House virus Flock House virus |
| Molecular weight | Theoretical: 113.996586 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MTLKVILGEH QITRTELLVG IATVSGCGAV VYCISKFWGY GAIAPYPQSG GNRVTRALQR AVIDKTKTPI ETRFYPLDSL RTVTPKRVA DNGHAVSGAV RDAARRLIDE SITAVGGSKF EVNPNPNSST GLRNHFHFAV GDLAQDFRND TPADDAFIVG V DVDYYVTE ...String: MTLKVILGEH QITRTELLVG IATVSGCGAV VYCISKFWGY GAIAPYPQSG GNRVTRALQR AVIDKTKTPI ETRFYPLDSL RTVTPKRVA DNGHAVSGAV RDAARRLIDE SITAVGGSKF EVNPNPNSST GLRNHFHFAV GDLAQDFRND TPADDAFIVG V DVDYYVTE PDVLLEHMRP VVLHTFNPKK VSGFDADSPF TIKNNLVEYK VSGGAAWVHP VWDWCEAGEF IASRVRTSWK EW FLQLPLR MIGLEKVGYH KIHHCRPWTD CPDRALVYTI PQYVIWRFNW IDTELHVRKL KRIEYQDETK PGWNRLEYVT DKN ELLVSI GREGEHAQIT IEKEKLDMLS GLSATQSVNA RLIGMGHKDP QYTSMIVQYY TGKKVVSPIS PTVYKPTMPR VHWP VTSDA DVPEVSARQY TLPIVSDCMM MPMIKRWETM SESIERRVTF VANDKKPSDR IAKIAETFVK LMNGPFKDLD PLSIE ETIE RLNKPSQQLQ LRAVFEMIGV KPRQLIESFN KNEPGMKSSR IISGFPDILF ILKVSRYTLA YSDIVLHAEH NEHWYY PGR NPTEIADGVC EFVSDCDAEV IETDFSNLDG RVSSWMQRNI AQKAMVQAFR PEYRDEIISF MDTIINCPAK AKRFGFR YE PGVGVKSGSP TTTPHNTQYN GCVEFTALTF EHPDAEPEDL FRLIGPKCGD DGLSRAIIQK SINRAAKCFG LELKVERY N PEIGLCFLSR VFVDPLATTT TIQDPLRTLR KLHLTTRDPT IPLADAACDR VEGYLCTDAL TPLISDYCKM VLRLYGPTA STEQVRNQRR SRNKEKPYWL TCDGSWPQHP QDAHLMKQVL IKRTAIDEDQ VDALIGRFAA MKDVWEKITH DSEESAAACT FDEDGVAPN SVDESLPLLN DAKQTRANPG TSRPHSNGGG SSHGNELPRR TEQRAQGPRQ PARLPKQGKT NGKSDGNITA G ETQRGGIP RGKGPRGGKT NTRRTPPKAG AQPQPSNNRK SRLEEELRRR LTE UniProtKB: RNA-directed RNA polymerase |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Support film - Film thickness: 2 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: INTEGRATING / Digitization - Dimensions - Width: 4096 pixel / Digitization - Dimensions - Height: 4096 pixel / Average electron dose: 100.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Protocol: FLEXIBLE FIT |
|---|---|
| Output model |  PDB-8fmb: |
 Movie
Movie Controller
Controller








 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)