[English] 日本語
 Yorodumi
Yorodumi- EMDB-29218: Nodavirus RNA replication crown from flock house virus-infected cells -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Nodavirus RNA replication crown from flock house virus-infected cells | |||||||||
 Map data Map data | Nodavirus RNA replication crown | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  Flock House virus Flock House virus | |||||||||
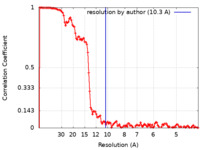
| Method | subtomogram averaging / cryo EM / Resolution: 10.3 Å | |||||||||
 Authors Authors | Zhan H / Unchwaniwala N / Rebolledo Viveros A / Pennington J / Horswill M / Broadberry R / Myers J / den Boon J / Grant T / Ahlquist P | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2023 Journal: Proc Natl Acad Sci U S A / Year: 2023Title: Nodavirus RNA replication crown architecture reveals proto-crown precursor and viral protein A conformational switching. Authors: Hong Zhan / Nuruddin Unchwaniwala / Andrea Rebolledo-Viveros / Janice Pennington / Mark Horswill / Roma Broadberry / Jonathan Myers / Johan A den Boon / Timothy Grant / Paul Ahlquist /  Abstract: Positive-strand RNA viruses replicate their genomes in virus-induced membrane vesicles, and the resulting RNA replication complexes are a major target for virus control. Nodavirus studies first ...Positive-strand RNA viruses replicate their genomes in virus-induced membrane vesicles, and the resulting RNA replication complexes are a major target for virus control. Nodavirus studies first revealed viral RNA replication proteins forming a 12-fold symmetric "crown" at the vesicle opening to the cytosol, an arrangement recently confirmed to extend to distantly related alphaviruses. Using cryoelectron microscopy (cryo-EM), we show that mature nodavirus crowns comprise two stacked 12-mer rings of multidomain viral RNA replication protein A. Each ring contains an ~19 nm circle of C-proximal polymerase domains, differentiated by strikingly diverged positions of N-proximal RNA capping/membrane binding domains. The lower ring is a "proto-crown" precursor that assembles prior to RNA template recruitment, RNA synthesis, and replication vesicle formation. In this proto-crown, the N-proximal segments interact to form a toroidal central floor, whose 3.1 Å resolution structure reveals many mechanistic details of the RNA capping/membrane binding domains. In the upper ring, cryo-EM fitting indicates that the N-proximal domains extend radially outside the polymerases, forming separated, membrane-binding "legs." The polymerase and N-proximal domains are connected by a long linker accommodating the conformational switch between the two rings and possibly also polymerase movements associated with RNA synthesis and nonsymmetric electron density in the lower center of mature crowns. The results reveal remarkable viral protein multifunctionality, conformational flexibility, and evolutionary plasticity and insights into (+)RNA virus replication and control. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_29218.map.gz emd_29218.map.gz | 129.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-29218-v30.xml emd-29218-v30.xml emd-29218.xml emd-29218.xml | 16.1 KB 16.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_29218_fsc.xml emd_29218_fsc.xml | 15.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_29218.png emd_29218.png | 108.2 KB | ||
| Others |  emd_29218_half_map_1.map.gz emd_29218_half_map_1.map.gz emd_29218_half_map_2.map.gz emd_29218_half_map_2.map.gz | 129.7 MB 129.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-29218 http://ftp.pdbj.org/pub/emdb/structures/EMD-29218 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-29218 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-29218 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_29218.map.gz / Format: CCP4 / Size: 139.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_29218.map.gz / Format: CCP4 / Size: 139.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Nodavirus RNA replication crown | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.156 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Nodavirus RNA replication crown unmasked half map1
| File | emd_29218_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Nodavirus RNA replication crown unmasked half map1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Nodavirus RNA replication crown unmasked half map2
| File | emd_29218_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Nodavirus RNA replication crown unmasked half map2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Flock House virus
| Entire | Name:  Flock House virus Flock House virus |
|---|---|
| Components |
|
-Supramolecule #1: Flock House virus
| Supramolecule | Name: Flock House virus / type: complex / ID: 1 / Chimera: Yes / Parent: 0 / Macromolecule list: all Details: Nodavirus RNA replication crown from flock house virus-infected Drosophila S2 cells |
|---|---|
| Source (natural) | Organism:  Flock House virus Flock House virus |
-Macromolecule #1: Flock House nodavirus protein A
| Macromolecule | Name: Flock House nodavirus protein A / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Flock House virus Flock House virus |
| Sequence | String: MTLKVILGEH QITRTELLVG IATVSGCGAV VYCISKFWGY GAIAPYPQSG GNRVTRALQR AVIDKTKTPI ETRFYPLDSL RTVTPKRVAD NGHAVSGAVR DAARRLIDES ITAVGGSKFE VNPNPNSSTG LRNHFHFAVG DLAQDFRNDT PADDAFIVGV DVDYYVTEPD ...String: MTLKVILGEH QITRTELLVG IATVSGCGAV VYCISKFWGY GAIAPYPQSG GNRVTRALQR AVIDKTKTPI ETRFYPLDSL RTVTPKRVAD NGHAVSGAVR DAARRLIDES ITAVGGSKFE VNPNPNSSTG LRNHFHFAVG DLAQDFRNDT PADDAFIVGV DVDYYVTEPD VLLEHMRPVV LHTFNPKKVS GFDADSPFTI KNNLVEYKVS GGAAWVHPVW DWCEAGEFIA SRVRTSWKEW FLQLPLRMIG LEKVGYHKIH HCRPWTDCPD RALVYTIPQY VIWRFNWIDT ELHVRKLKRI EYQDETKPGW NRLEYVTDKN ELLVSIGREG EHAQITIEKE KLDMLSGLSA TQSVNARLIG MGHKDPQYTS MIVQYYTGKK VVSPISPTVY KPTMPRVHWP VTSDADVPEV SARQYTLPIV SDCMMMPMIK RWETMSESIE RRVTFVANDK KPSDRIAKIA ETFVKLMNGP FKDLDPLSIE ETIERLNKPS QQLQLRAVFE MIGVKPRQLI ESFNKNEPGM KSSRIISGFP DILFILKVSR YTLAYSDIVL HAEHNEHWYY PGRNPTEIAD GVCEFVSDCD AEVIETDFSN LDGRVSSWMQ RNIAQKAMVQ AFRPEYRDEI ISFMDTIINC PAKAKRFGFR YEPGVGVKSG SPTTTPHNTQ YNGCVEFTAL TFEHPDAEPE DLFRLIGPKC GDDGLSRAII QKSINRAAKC FGLELKVERY NPEIGLCFLS RVFVDPLATT TTIQDPLRTL RKLHLTTRDP TIPLADAACD RVEGYLCTDA LTPLISDYCK MVLRLYGPTA STEQVRNQRR SRNKEKPYWL TCDGSWPQHP QDAHLMKQVL IKRTAIDEDQ VDALIGRFAA MKDVWEKITH DSEESAAACT FDEDGVAPNS VDESLPMLND AKQTRANPGT SRPHSNGGGS SHGNELPRRT EQRAQGPRQP ARLPKQGKTN GKSDGNITAG ETQRGGIPRG KGPRGGKTNT RRTPPKAGAQ PQPSNNRK |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | subtomogram averaging |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Grid | Model: Quantifoil R2/2 / Material: COPPER / Mesh: 200 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 60 sec. |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 295 K / Instrument: FEI VITROBOT MARK III |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Software | Name: SerialEM (ver. 3.8) |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 4.86 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 3.5 µm / Nominal defocus min: 3.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)