登録情報 データベース : EMDB / ID : EMD-29213タイトル Cryo-EM structure of E. coli RNA polymerase Elongation complex in the Transcription-Translation Complex (RNAP in an anti-swiveled conformation) Cryo-EM structure of E. coli RNA polymerase Elongation complex (in the Transcription-Translation Complex) harboring a terminal mismatch 複合体 : Cryo-EM structure of E. coli RNA polymerase Elongation complex (in the Transcription-Translation Complex) harboring a terminal mismatchタンパク質・ペプチド : DNA-directed RNA polymerase subunit alphaタンパク質・ペプチド : DNA-directed RNA polymerase subunit betaタンパク質・ペプチド : DNA-directed RNA polymerase subunit beta'DNA : Non-template DNADNA : Template DNARNA : RNAリガンド : ZINC IONリガンド : MAGNESIUM ION / / / / 機能・相同性 分子機能 ドメイン・相同性 構成要素
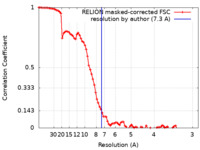
/ / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / 生物種 Escherichia coli K-12 (大腸菌) / 手法 / / 解像度 : 7.3 Å Florez Ariza A / Wee L / Tong A / Canari C / Grob P / Nogales E / Bustamante C 資金援助 Organization Grant number 国 Howard Hughes Medical Institute (HHMI) National Institutes of Health/National Center for Research Resources (NIH/NCRR)
ジャーナル : Cell / 年 : 2023タイトル : A trailing ribosome speeds up RNA polymerase at the expense of transcript fidelity via force and allostery.著者 : Liang Meng Wee / Alexander B Tong / Alfredo Jose Florez Ariza / Cristhian Cañari-Chumpitaz / Patricia Grob / Eva Nogales / Carlos J Bustamante / 要旨 : In prokaryotes, translation can occur on mRNA that is being transcribed in a process called coupling. How the ribosome affects the RNA polymerase (RNAP) during coupling is not well understood. Here, ... In prokaryotes, translation can occur on mRNA that is being transcribed in a process called coupling. How the ribosome affects the RNA polymerase (RNAP) during coupling is not well understood. Here, we reconstituted the E. coli coupling system and demonstrated that the ribosome can prevent pausing and termination of RNAP and double the overall transcription rate at the expense of fidelity. Moreover, we monitored single RNAPs coupled to ribosomes and show that coupling increases the pause-free velocity of the polymerase and that a mechanical assisting force is sufficient to explain the majority of the effects of coupling. Also, by cryo-EM, we observed that RNAPs with a terminal mismatch adopt a backtracked conformation, while a coupled ribosome allosterically induces these polymerases toward a catalytically active anti-swiveled state. Finally, we demonstrate that prolonged RNAP pausing is detrimental to cell viability, which could be prevented by polymerase reactivation through a coupled ribosome. 履歴 登録 2022年12月17日 - ヘッダ(付随情報) 公開 2023年3月29日 - マップ公開 2023年3月29日 - 更新 2025年5月14日 - 現状 2025年5月14日 処理サイト : RCSB / 状態 : 公開
すべて表示 表示を減らす
 データを開く
データを開く 基本情報
基本情報
 マップデータ
マップデータ 試料
試料 キーワード
キーワード 機能・相同性情報
機能・相同性情報
 Escherichia phage Lambda (λファージ)
Escherichia phage Lambda (λファージ) データ登録者
データ登録者 米国, 2件
米国, 2件  引用
引用 ジャーナル: Cell / 年: 2023
ジャーナル: Cell / 年: 2023
 構造の表示
構造の表示 ダウンロードとリンク
ダウンロードとリンク emd_29213.map.gz
emd_29213.map.gz EMDBマップデータ形式
EMDBマップデータ形式 emd-29213-v30.xml
emd-29213-v30.xml emd-29213.xml
emd-29213.xml EMDBヘッダ
EMDBヘッダ emd_29213_fsc.xml
emd_29213_fsc.xml FSCデータファイル
FSCデータファイル emd_29213.png
emd_29213.png emd_29213_msk_1.map
emd_29213_msk_1.map マスクマップ
マスクマップ emd-29213.cif.gz
emd-29213.cif.gz emd_29213_half_map_1.map.gz
emd_29213_half_map_1.map.gz emd_29213_half_map_2.map.gz
emd_29213_half_map_2.map.gz http://ftp.pdbj.org/pub/emdb/structures/EMD-29213
http://ftp.pdbj.org/pub/emdb/structures/EMD-29213 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-29213
ftp://ftp.pdbj.org/pub/emdb/structures/EMD-29213 emd_29213_validation.pdf.gz
emd_29213_validation.pdf.gz EMDB検証レポート
EMDB検証レポート emd_29213_full_validation.pdf.gz
emd_29213_full_validation.pdf.gz emd_29213_validation.xml.gz
emd_29213_validation.xml.gz emd_29213_validation.cif.gz
emd_29213_validation.cif.gz https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-29213
https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-29213 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-29213
ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-29213


 F&H 検索
F&H 検索 リンク
リンク EMDB (EBI/PDBe) /
EMDB (EBI/PDBe) /  EMDataResource
EMDataResource マップ
マップ ダウンロード / ファイル: emd_29213.map.gz / 形式: CCP4 / 大きさ: 52.7 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES)
ダウンロード / ファイル: emd_29213.map.gz / 形式: CCP4 / 大きさ: 52.7 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) emd_29213_msk_1.map
emd_29213_msk_1.map 試料の構成要素
試料の構成要素





 Escherichia phage Lambda (λファージ)
Escherichia phage Lambda (λファージ) Escherichia phage Lambda (λファージ)
Escherichia phage Lambda (λファージ) Escherichia phage Lambda (λファージ)
Escherichia phage Lambda (λファージ) 解析
解析 試料調製
試料調製 電子顕微鏡法
電子顕微鏡法 FIELD EMISSION GUN
FIELD EMISSION GUN
 ムービー
ムービー コントローラー
コントローラー










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)