[English] 日本語
 Yorodumi
Yorodumi- EMDB-29115: Human nucleolar pre-60S ribosomal subunit (State H) - Overall map -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Human nucleolar pre-60S ribosomal subunit (State H) - Overall map | |||||||||
 Map data Map data | Local resolution filtered map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Pre-60S ribosomal subunit / Assembly intermediate / Ribosome / Nucleoprotein complex | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
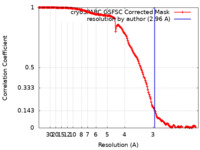
| Method | single particle reconstruction / cryo EM / Resolution: 2.96 Å | |||||||||
 Authors Authors | Vanden Broeck A / Klinge S | |||||||||
| Funding support | European Union,  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Science / Year: 2023 Journal: Science / Year: 2023Title: Principles of human pre-60 biogenesis. Authors: Arnaud Vanden Broeck / Sebastian Klinge /  Abstract: During the early stages of human large ribosomal subunit (60) biogenesis, an ensemble of assembly factors establishes and fine-tunes the essential RNA functional centers of pre-60 particles by an ...During the early stages of human large ribosomal subunit (60) biogenesis, an ensemble of assembly factors establishes and fine-tunes the essential RNA functional centers of pre-60 particles by an unknown mechanism. Here, we report a series of cryo-electron microscopy structures of human nucleolar and nuclear pre-60 assembly intermediates at resolutions of 2.5 to 3.2 angstroms. These structures show how protein interaction hubs tether assembly factor complexes to nucleolar particles and how guanosine triphosphatases and adenosine triphosphatase couple irreversible nucleotide hydrolysis steps to the installation of functional centers. Nuclear stages highlight how a conserved RNA-processing complex, the rixosome, couples large-scale RNA conformational changes with pre-ribosomal RNA processing by the RNA degradation machinery. Our ensemble of human pre-60 particles provides a rich foundation with which to elucidate the molecular principles of ribosome formation. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_29115.map.gz emd_29115.map.gz | 40.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-29115-v30.xml emd-29115-v30.xml emd-29115.xml emd-29115.xml | 25.2 KB 25.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_29115_fsc.xml emd_29115_fsc.xml | 15.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_29115.png emd_29115.png | 125.9 KB | ||
| Masks |  emd_29115_msk_1.map emd_29115_msk_1.map | 421.9 MB |  Mask map Mask map | |
| Others |  emd_29115_additional_1.map.gz emd_29115_additional_1.map.gz emd_29115_additional_2.map.gz emd_29115_additional_2.map.gz emd_29115_half_map_1.map.gz emd_29115_half_map_1.map.gz emd_29115_half_map_2.map.gz emd_29115_half_map_2.map.gz | 217.8 MB 332.4 MB 391.2 MB 391.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-29115 http://ftp.pdbj.org/pub/emdb/structures/EMD-29115 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-29115 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-29115 | HTTPS FTP |
-Validation report
| Summary document |  emd_29115_validation.pdf.gz emd_29115_validation.pdf.gz | 1.1 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_29115_full_validation.pdf.gz emd_29115_full_validation.pdf.gz | 1.1 MB | Display | |
| Data in XML |  emd_29115_validation.xml.gz emd_29115_validation.xml.gz | 25 KB | Display | |
| Data in CIF |  emd_29115_validation.cif.gz emd_29115_validation.cif.gz | 32.7 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-29115 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-29115 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-29115 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-29115 | HTTPS FTP |
-Related structure data
| Related structure data |  8fkpC  8fkqC  8fkrC  8fksC  8fktC  8fkuC  8fkvC  8fkwC  8fkxC  8fkyC  8fkzC  8fl0C  8fl2C  8fl3C  8fl4C  8fl6C  8fl7C  8fl9C  8flaC  8flbC  8flcC  8fldC  8fleC  8flfC C: citing same article ( |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_29115.map.gz / Format: CCP4 / Size: 421.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_29115.map.gz / Format: CCP4 / Size: 421.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Local resolution filtered map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.072 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_29115_msk_1.map emd_29115_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: cryoSPARC sharpened map
| File | emd_29115_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | cryoSPARC sharpened map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Phenix auto-sharpened map
| File | emd_29115_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Phenix auto-sharpened map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_29115_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_29115_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Human nucleolar pre-60S ribosomal subunit (State H)
| Entire | Name: Human nucleolar pre-60S ribosomal subunit (State H) |
|---|---|
| Components |
|
-Supramolecule #1: Human nucleolar pre-60S ribosomal subunit (State H)
| Supramolecule | Name: Human nucleolar pre-60S ribosomal subunit (State H) / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#47 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) / Strain: HEK293F Homo sapiens (human) / Strain: HEK293F |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.6 |
|---|---|
| Grid | Model: Quantifoil R3.5/1 / Material: GOLD / Mesh: 400 / Support film - Material: CARBON / Support film - topology: CONTINUOUS / Support film - Film thickness: 2 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 283 K / Instrument: FEI VITROBOT MARK IV Details: Four applications with manual blotting before last blotting with the vitrobot.. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Number grids imaged: 4 / Number real images: 172699 / Average exposure time: 2.0 sec. / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.5 µm / Nominal magnification: 64000 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller





























































































































 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)