+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of human NCC | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Membrane transporter / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationDefective SLC12A3 causes Gitelman syndrome (GS) / sodium:chloride symporter activity / sodium:potassium:chloride symporter activity / Cation-coupled Chloride cotransporters / sodium ion homeostasis / chloride ion homeostasis / renal sodium ion absorption / response to salt / potassium ion homeostasis / response to aldosterone ...Defective SLC12A3 causes Gitelman syndrome (GS) / sodium:chloride symporter activity / sodium:potassium:chloride symporter activity / Cation-coupled Chloride cotransporters / sodium ion homeostasis / chloride ion homeostasis / renal sodium ion absorption / response to salt / potassium ion homeostasis / response to aldosterone / cell volume homeostasis / sodium ion transport / potassium ion import across plasma membrane / monoatomic ion transport / chloride transmembrane transport / sodium ion transmembrane transport / apical plasma membrane / extracellular exosome / ATP binding / membrane / plasma membrane / cytosol Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
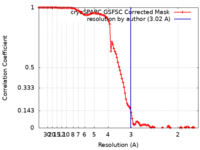
| Method | single particle reconstruction / cryo EM / Resolution: 3.02 Å | |||||||||
 Authors Authors | Zhang J / Fan M / Feng L | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: Nature / Year: 2023 Journal: Nature / Year: 2023Title: Structure and thiazide inhibition mechanism of the human Na-Cl cotransporter. Authors: Minrui Fan / Jianxiu Zhang / Chien-Ling Lee / Jinru Zhang / Liang Feng /  Abstract: The sodium-chloride cotransporter (NCC) is critical for kidney physiology. The NCC has a major role in salt reabsorption in the distal convoluted tubule of the nephron, and mutations in the NCC cause ...The sodium-chloride cotransporter (NCC) is critical for kidney physiology. The NCC has a major role in salt reabsorption in the distal convoluted tubule of the nephron, and mutations in the NCC cause the salt-wasting disease Gitelman syndrome. As a key player in salt handling, the NCC regulates blood pressure and is the target of thiazide diuretics, which have been widely prescribed as first-line medications to treat hypertension for more than 60 years. Here we determined the structures of human NCC alone and in complex with a commonly used thiazide diuretic using cryo-electron microscopy. These structures, together with functional studies, reveal major conformational states of the NCC and an intriguing regulatory mechanism. They also illuminate how thiazide diuretics specifically interact with the NCC and inhibit its transport function. Our results provide critical insights for understanding the Na-Cl cotransport mechanism of the NCC, and they establish a framework for future drug design and for interpreting disease-related mutations. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_29103.map.gz emd_29103.map.gz | 64.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-29103-v30.xml emd-29103-v30.xml emd-29103.xml emd-29103.xml | 18.7 KB 18.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_29103_fsc.xml emd_29103_fsc.xml | 10.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_29103.png emd_29103.png | 81.2 KB | ||
| Masks |  emd_29103_msk_1.map emd_29103_msk_1.map | 125 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-29103.cif.gz emd-29103.cif.gz | 6.6 KB | ||
| Others |  emd_29103_half_map_1.map.gz emd_29103_half_map_1.map.gz emd_29103_half_map_2.map.gz emd_29103_half_map_2.map.gz | 116 MB 116 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-29103 http://ftp.pdbj.org/pub/emdb/structures/EMD-29103 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-29103 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-29103 | HTTPS FTP |
-Related structure data
| Related structure data |  8fhtMC  8fhnC  8fhoC  8fhpC  8fhqC  8fhrC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_29103.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_29103.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.86 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_29103_msk_1.map emd_29103_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_29103_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_29103_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : NCC
| Entire | Name: NCC |
|---|---|
| Components |
|
-Supramolecule #1: NCC
| Supramolecule | Name: NCC / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Solute carrier family 12 member 3
| Macromolecule | Name: Solute carrier family 12 member 3 / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 113.279719 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MAELPTTETP GDATLCSGRF TISTLLSSDE PSPPAAYDSS HPSHLTHSST FCMRDFGYND IDVVPTYEHY ANDTQPGEPR KVRPTLADL HSFLKQEGRH LHALAFDSRP SHEMTDGLVE GEAGTSSEKN PEEPVRFGWV NGVMIRCMLN IWGVILYLRL P WITAQAGI ...String: MAELPTTETP GDATLCSGRF TISTLLSSDE PSPPAAYDSS HPSHLTHSST FCMRDFGYND IDVVPTYEHY ANDTQPGEPR KVRPTLADL HSFLKQEGRH LHALAFDSRP SHEMTDGLVE GEAGTSSEKN PEEPVRFGWV NGVMIRCMLN IWGVILYLRL P WITAQAGI VLTWIIILLS VTVTSITGLS ISAISTNGKV KSGGTYFLIS RSLGPELGGS IGLIFAFANA VGVAMHTVGF AE TVRDLLQ EYGAPIVDPI NDIRIIGVVS VTVLLAISLA GMEWESKAQV LFFLVIMVSF ANYLVGTLIP PSEDKASKGF FSY RADIFV QNLVPDWRGP DGTFFGMFSI FFPSATGILA GANISGDLKD PAIAIPKGTL MAIFWTTISY LAISATIGSC VVRD ASGVL NDTVTPGWGA CEGLACSYGW NFTECTQQHS CHYGLINYYQ TMSMVSGFAP LITAGIFGAT LSSALACLVS AAKVF QCLC EDQLYPLIGF FGKGYGKNKE PVRGYLLAYA IAVAFIIIAE LNTIAPIISN FFLCSYALIN FSCFHASITN SPGWRP SFQ YYNKWAALFG AIISVVIMFL LTWWAALIAI GVVLFLLLYV IYKKPEVNWG SSVQAGSYNL ALSYSVGLNE VEDHIKN YR PQCLVLTGPP NFRPALVDFV GTFTRNLSLM ICGHVLIGPH KQRMPELQLI ANGHTKWLNK RKIKAFYSDV IAEDLRRG V QILMQAAGLG RMKPNILVVG FKKNWQSAHP ATVEDYIGIL HDAFDFNYGV CVMRMREGLN VSKMMQAHIN PVFDPAEDG KEASARVDPK ALVKEEQATT IFQSEQGKKT IDIYWLFDDG GLTLLIPYLL GRKRRWSKCK IRVFVGGQIN RMDQERKAII SLLSKFRLG FHEVHILPDI NQNPRAEHTK RFEDMIAPFR LNDGFKDEAT VNEMRRDCPW KISDEEITKN RVKSLRQVRL N EIVLDYSR DAALIVITLP IGRKGKCPSS LYMAWLETLS QDLRPPVILI RGNQENVLTF YCQ UniProtKB: Solute carrier family 12 member 3 |
-Macromolecule #2: SODIUM ION
| Macromolecule | Name: SODIUM ION / type: ligand / ID: 2 / Number of copies: 2 |
|---|---|
| Molecular weight | Theoretical: 22.99 Da |
-Macromolecule #3: CHLORIDE ION
| Macromolecule | Name: CHLORIDE ION / type: ligand / ID: 3 / Number of copies: 2 / Formula: CL |
|---|---|
| Molecular weight | Theoretical: 35.453 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 12 mg/mL |
|---|---|
| Buffer | pH: 7.4 |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.2 µm / Nominal defocus min: 1.2 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)