+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of human NCC (class 3-1) | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Membrane transporter / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationswim bladder inflation / Cation-coupled Chloride cotransporters / Defective SLC12A3 causes Gitelman syndrome (GS) / sodium:chloride symporter activity / chloride:monoatomic cation symporter activity / sodium:potassium:chloride symporter activity / Cation-coupled Chloride cotransporters / ammonium transmembrane transport / sodium ion homeostasis / ammonium channel activity ...swim bladder inflation / Cation-coupled Chloride cotransporters / Defective SLC12A3 causes Gitelman syndrome (GS) / sodium:chloride symporter activity / chloride:monoatomic cation symporter activity / sodium:potassium:chloride symporter activity / Cation-coupled Chloride cotransporters / ammonium transmembrane transport / sodium ion homeostasis / ammonium channel activity / chloride ion homeostasis / renal sodium ion absorption / ear development / response to salt / potassium ion homeostasis / response to aldosterone / cell volume homeostasis / inner ear morphogenesis / sodium ion transport / potassium ion import across plasma membrane / monoatomic ion transport / chloride transmembrane transport / sodium ion transmembrane transport / basolateral plasma membrane / apical plasma membrane / extracellular exosome / ATP binding / metal ion binding / identical protein binding / membrane / plasma membrane / cytosol Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
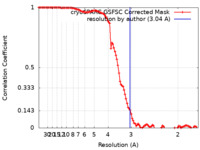
| Method | single particle reconstruction / cryo EM / Resolution: 3.04 Å | |||||||||
 Authors Authors | Zhang J / Fan M / Feng L | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: Nature / Year: 2023 Journal: Nature / Year: 2023Title: Structure and thiazide inhibition mechanism of the human Na-Cl cotransporter. Authors: Minrui Fan / Jianxiu Zhang / Chien-Ling Lee / Jinru Zhang / Liang Feng /  Abstract: The sodium-chloride cotransporter (NCC) is critical for kidney physiology. The NCC has a major role in salt reabsorption in the distal convoluted tubule of the nephron, and mutations in the NCC cause ...The sodium-chloride cotransporter (NCC) is critical for kidney physiology. The NCC has a major role in salt reabsorption in the distal convoluted tubule of the nephron, and mutations in the NCC cause the salt-wasting disease Gitelman syndrome. As a key player in salt handling, the NCC regulates blood pressure and is the target of thiazide diuretics, which have been widely prescribed as first-line medications to treat hypertension for more than 60 years. Here we determined the structures of human NCC alone and in complex with a commonly used thiazide diuretic using cryo-electron microscopy. These structures, together with functional studies, reveal major conformational states of the NCC and an intriguing regulatory mechanism. They also illuminate how thiazide diuretics specifically interact with the NCC and inhibit its transport function. Our results provide critical insights for understanding the Na-Cl cotransport mechanism of the NCC, and they establish a framework for future drug design and for interpreting disease-related mutations. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_29098.map.gz emd_29098.map.gz | 64.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-29098-v30.xml emd-29098-v30.xml emd-29098.xml emd-29098.xml | 18.9 KB 18.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_29098_fsc.xml emd_29098_fsc.xml | 10.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_29098.png emd_29098.png | 89.3 KB | ||
| Masks |  emd_29098_msk_1.map emd_29098_msk_1.map | 125 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-29098.cif.gz emd-29098.cif.gz | 6.9 KB | ||
| Others |  emd_29098_half_map_1.map.gz emd_29098_half_map_1.map.gz emd_29098_half_map_2.map.gz emd_29098_half_map_2.map.gz | 115.8 MB 115.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-29098 http://ftp.pdbj.org/pub/emdb/structures/EMD-29098 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-29098 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-29098 | HTTPS FTP |
-Related structure data
| Related structure data |  8fhpMC  8fhnC  8fhoC  8fhqC  8fhrC  8fhtC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_29098.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_29098.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.86 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_29098_msk_1.map emd_29098_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_29098_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_29098_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : NCC-polythiazide complex
| Entire | Name: NCC-polythiazide complex |
|---|---|
| Components |
|
-Supramolecule #1: NCC-polythiazide complex
| Supramolecule | Name: NCC-polythiazide complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Solute carrier family 12 member 2,Solute carrier family 12 member...
| Macromolecule | Name: Solute carrier family 12 member 2,Solute carrier family 12 member 3 chimera type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 120.372164 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MSASPPISAG DYLSAPEPDA LKPAGPTPSQ SRFQVDLVTE SAGDGETTVG FDSSPPEYVA EPPPDGLRDS VSGGEKAKGR FRVVNFAAS SPDAAPAETA QNGDTVMSEG SLHSSTGGQQ HHHYDTHTNT YYLRTFGHNT IDAVPKIDFY RQTAAPLGEK L IRPTLSEL ...String: MSASPPISAG DYLSAPEPDA LKPAGPTPSQ SRFQVDLVTE SAGDGETTVG FDSSPPEYVA EPPPDGLRDS VSGGEKAKGR FRVVNFAAS SPDAAPAETA QNGDTVMSEG SLHSSTGGQQ HHHYDTHTNT YYLRTFGHNT IDAVPKIDFY RQTAAPLGEK L IRPTLSEL HDELDKEPFE DGFANGEELT PAEESAAKDV SESKEPVRFG WVKGVMIRCM LNIWGVILYL RLPWITAQAG IV LTWIIIL LSVTVTSITG LSISAISTNG KVKSGGTYFL ISRSLGPELG GSIGLIFAFA NAVGVAMHTV GFAATVRDLL QEY GAPIVD PINDIRIIGV VSVTVLLAIS LAGMEWESKA QVLFFLVIMV SFANYLVGTL IPPSEDKASK GFFSYRADIF VQNL VPDWR GPDGTFFGMF SIFFPSATGI LAGANISGDL KDPAIAIPKG TLMAIFWTTI SYLAISATIG SCVVRDASGV LNDTV TPGW GACEGLACSY GWNFTECTQQ HSCHYGLINY YQTMSMVSGF APLITAGIFG ATLSSALACL VSAAKVFQCL CEDQLY PLI GFFGKGYGKN KEPVRGYLLA YAIAVAFIII AELNTIAPII SNFFLCSYAL INFSCFHASI TNSPGWRPSF QYYNKWA AL FGAIISVVIM FLLTWWAALI AIGVVLFLLL YVIYKKPEVN WGSSVQAGSY NLALSYSVGL NEVEDHIKNY RPQCLVLT G PPNFRPALVD FVGTFTRNLS LMICGHVLIG PHKQRMPELQ LIANGHTKWL NKRKIKAFYS DVIAEDLRRG VQILMQAAG LGRMKPNILV VGFKKNWQSA HPATVEDYIG ILHDAFDFNY GVCVMRMREG LNVSKMMQAH INPVFDPAED GKEASARVDP KALVKEEQA TTIFQSEQGK KTIDIYWLFD DGGLTLLIPY LLGRKRRWSK CKIRVFVGGQ INRMDQERKA IISLLSKFRL G FHEVHILP DINQNPRAEH TKRFEDMIAP FRLNDGFKDE ATVNEMRRDC PWKISDEEIT KNRVKSLRQV RLNEIVLDYS RD AALIVIT LPIGRKGKCP SSLYMAWLET LSQDLRPPVI LIRGNQENVL TFYCQ UniProtKB: Solute carrier family 12 member 2, Solute carrier family 12 member 3 |
-Macromolecule #2: Polythiazide
| Macromolecule | Name: Polythiazide / type: ligand / ID: 2 / Number of copies: 2 / Formula: XZF |
|---|---|
| Molecular weight | Theoretical: 439.882 Da |
| Chemical component information | 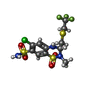 ChemComp-XZF: |
-Macromolecule #3: SODIUM ION
| Macromolecule | Name: SODIUM ION / type: ligand / ID: 3 / Number of copies: 2 |
|---|---|
| Molecular weight | Theoretical: 22.99 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 12 mg/mL |
|---|---|
| Buffer | pH: 7.4 |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.2 µm / Nominal defocus min: 1.2 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)