+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of coagulation factor V short | ||||||||||||
 Map data Map data | Primary Map | ||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | BLOOD CLOTTING | ||||||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||||||||
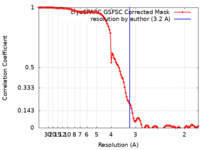
| Method | single particle reconstruction / cryo EM / Resolution: 3.2 Å | ||||||||||||
 Authors Authors | Mohammed BM / Pelc LA / Rau MJ / Di Cera E | ||||||||||||
| Funding support |  United States, 3 items United States, 3 items
| ||||||||||||
 Citation Citation |  Journal: Blood / Year: 2023 Journal: Blood / Year: 2023Title: Cryo-EM structure of coagulation factor V short. Authors: Bassem M Mohammed / Leslie A Pelc / Michael J Rau / Enrico Di Cera /  Abstract: Coagulation factor V (fV) is the precursor of activated fV (fVa), an essential component of the prothrombinase complex required for the rapid activation of prothrombin in the penultimate step of the ...Coagulation factor V (fV) is the precursor of activated fV (fVa), an essential component of the prothrombinase complex required for the rapid activation of prothrombin in the penultimate step of the coagulation cascade. In addition, fV regulates the tissue factor pathway inhibitor α (TFPIα) and protein C pathways that inhibit the coagulation response. A recent cryogenic electron microscopy (cryo-EM) structure of fV has revealed the architecture of its A1-A2-B-A3-C1-C2 assembly but left the mechanism that keeps fV in its inactive state unresolved because of an intrinsic disorder in the B domain. A splice variant of fV, fV short, carries a large deletion of the B domain that produces constitutive fVa-like activity and unmasks epitopes for the binding of TFPIα. The cryo-EM structure of fV short was solved at 3.2 Å resolution and revealed the arrangement of the entire A1-A2-B-A3-C1-C2 assembly. The shorter B domain stretches across the entire width of the protein, making contacts with the A1, A2, and A3 domains but suspended over the C1 and C2 domains. In the portion distal to the splice site, several hydrophobic clusters and acidic residues provide a potential binding site for the basic C-terminal end of TFPIα. In fV, these epitopes may bind intramolecularly to the basic region of the B domain. The cryo-EM structure reported in this study advances our understanding of the mechanism that keeps fV in its inactive state, provides new targets for mutagenesis and facilitates future structural analysis of fV short in complex with TFPIα, protein S, and fXa. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_29009.map.gz emd_29009.map.gz | 72.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-29009-v30.xml emd-29009-v30.xml emd-29009.xml emd-29009.xml | 17.9 KB 17.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_29009_fsc.xml emd_29009_fsc.xml | 11.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_29009.png emd_29009.png | 70 KB | ||
| Masks |  emd_29009_msk_1.map emd_29009_msk_1.map | 144.7 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-29009.cif.gz emd-29009.cif.gz | 6 KB | ||
| Others |  emd_29009_half_map_1.map.gz emd_29009_half_map_1.map.gz emd_29009_half_map_2.map.gz emd_29009_half_map_2.map.gz | 134.1 MB 134.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-29009 http://ftp.pdbj.org/pub/emdb/structures/EMD-29009 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-29009 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-29009 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_29009.map.gz / Format: CCP4 / Size: 144.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_29009.map.gz / Format: CCP4 / Size: 144.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Primary Map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.9 Å | ||||||||||||||||||||||||||||||||||||
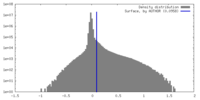
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_29009_msk_1.map emd_29009_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half Map 1
| File | emd_29009_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half Map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half Map 2
| File | emd_29009_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half Map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Recombinantly expressed coagulation factor V short full length wi...
| Entire | Name: Recombinantly expressed coagulation factor V short full length with a c-terminus HPC4 tag |
|---|---|
| Components |
|
-Supramolecule #1: Recombinantly expressed coagulation factor V short full length wi...
| Supramolecule | Name: Recombinantly expressed coagulation factor V short full length with a c-terminus HPC4 tag type: tissue / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) / Organ: Liver / Tissue: Blood Homo sapiens (human) / Organ: Liver / Tissue: Blood |
-Macromolecule #1: Coagulation Factor V short
| Macromolecule | Name: Coagulation Factor V short / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) / Organ: Liver / Tissue: Blood Homo sapiens (human) / Organ: Liver / Tissue: Blood |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: AQLRQFYVAA QGISWSYRPE PTNSSLNLSV TSFKKIVYRE YEPYFKKEKP QSTISGLLGP TLYAEVGDII KVHFKNKADK PLSIHPQGIR YSKLSEGASY LDHTFPAEKM DDAVAPGREY TYEWSISEDS GPTHDDPPCL THIYYSHENL IEDFNSGLIG PLLICKKGTL ...String: AQLRQFYVAA QGISWSYRPE PTNSSLNLSV TSFKKIVYRE YEPYFKKEKP QSTISGLLGP TLYAEVGDII KVHFKNKADK PLSIHPQGIR YSKLSEGASY LDHTFPAEKM DDAVAPGREY TYEWSISEDS GPTHDDPPCL THIYYSHENL IEDFNSGLIG PLLICKKGTL TEGGTQKTFD KQIVLLFAVF DESKSWSQSS SLMYTVNGYV NGTMPDITVC AHDHISWHLL GMSSGPELFS IHFNGQVLEQ NHHKVSAITL VSATSTTANM TVGPEGKWII SSLTPKHLQA GMQAYIDIKN CPKKTRNLKK ITREQRRHMK RWEYFIAAEE VIWDYAPVIP ANMDKKYRSQ HLDNFSNQIG KHYKKVMYTQ YEDESFTKHT VNPNMKEDGI LGPIIRAQVR DTLKIVFKNM ASRPYSIYPH GVTFSPYEDE VNSSFTSGRN NTMIRAVQPG ETYTYKWNIL EFDEPTENDA QCLTRPYYSD VDIMRDIASG LIGLLLICKS RSLDRRGIQR AADIEQQAVF AVFDENKSWY LEDNINKFCE NPDEVKRDDP KFYESNIMST INGYVPESIT TLGFCFDDTV QWHFCSVGTQ NEILTIHFTG HSFIYGKRHE DTLTLFPMRG ESVTVTMDNV GTWMLTSMNS SPRSKKLRLK FRDVKCIPDD DEDSYEIFEP PESTVMATRK MHDRLEPEDE ESDADYDYQN RLAAALGIRS FRNSSLNQEE EEFNLTALAL ENGTEFVSSN TDIIVGSNYS SPSNILGQMP SPSSPTLNDT FLSKEFNPLV IVGLSKDGTD YIEIIPKEEV QSSEDDYAEI DYVPYDDPYK TDVRTNINSS RDPDNIAAWY LRSNNGNRRN YYIAAEEISW DYSEFVQRET DIEDSDDIPE DTTYKKVVFR KYLDSTFTKR DPRGEYEEHL GILGPIIRAE VDDVIQVRFK NLASRPYSLH AHGLSYEKSS EGKTYEDDSP EWFKEDNAVQ PNSSYTYVWH ATERSGPESP GSACRAWAYY SAVNPEKDIH SGLIGPLLIC QKGILHKDSN MPVDMREFVL LFMTFDEKKS WYYEKKSRSS WRLTSSEMKK SHEFHAINGM IYSLPGLKMY EQEWVRLHLL NIGGSQDIHV VHFHGQTLLE NGNKQHQLGV WPLLPGSFKT LEMKASKPGW WLLNTEVGEN QRAGMQTPFL IMDRDCRMPM GLSTGIISDS QIKASEFLGY WEPRLARLNN GGSYNAWSVE KLAAEFASKP WIQVDMQKEV IITGIQTQGA KHYLKSCYTT EFYVAYSSNQ INWQIFKGNS TRNVMYFNGN SDASTIKENQ FDPPIVARYI RISPTRAYNR PTLRLELQGC EVNGCSTPLG MENGKIENKQ ITASSFKKSW WGDYWEPFRA RLNAQGRVNA WQAKANNNKQ WLEIDLLKIK KITAIITQGC KSLSSEMYVK SYTIHYSEQG VEWKPYRLKS SMVDKIFEGN TNTKGHVKNF FNPPIISRFI RVIPKTWNQS ITLRLELFGC DIYEDQVDPR LIDGK |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.1 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.4 Component:
| ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Instrument: FEI VITROBOT MARK IV / Details: 2 second blot 20 second wait time. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Spherical aberration corrector: Cs corrector |
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Detector mode: COUNTING / Digitization - Dimensions - Width: 4096 pixel / Digitization - Dimensions - Height: 4096 pixel / Digitization - Frames/image: 1-46 / Number grids imaged: 1 / Number real images: 3316 / Average exposure time: 9.36 sec. / Average electron dose: 55.09 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 0.01 mm / Nominal defocus max: 2.4 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 75000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: AB INITIO MODEL |
|---|
 Movie
Movie Controller
Controller








 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)