+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Gi bound delta-opioid receptor in complex with deltorphin | ||||||||||||
 Map data Map data | |||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | delta opioid receptor / G protein coupled receptor / deltorphin / SIGNALING PROTEIN | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationopioid peptide activity / G protein-coupled enkephalin receptor activity / spine apparatus / G protein-coupled opioid receptor activity / G protein-coupled opioid receptor signaling pathway / receptor serine/threonine kinase binding / cellular response to toxic substance / G-protein activation / Activation of the phototransduction cascade / Glucagon-type ligand receptors ...opioid peptide activity / G protein-coupled enkephalin receptor activity / spine apparatus / G protein-coupled opioid receptor activity / G protein-coupled opioid receptor signaling pathway / receptor serine/threonine kinase binding / cellular response to toxic substance / G-protein activation / Activation of the phototransduction cascade / Glucagon-type ligand receptors / Thromboxane signalling through TP receptor / Sensory perception of sweet, bitter, and umami (glutamate) taste / G beta:gamma signalling through PI3Kgamma / G beta:gamma signalling through CDC42 / Cooperation of PDCL (PhLP1) and TRiC/CCT in G-protein beta folding / Activation of G protein gated Potassium channels / Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits / Ca2+ pathway / G alpha (z) signalling events / High laminar flow shear stress activates signaling by PIEZO1 and PECAM1:CDH5:KDR in endothelial cells / Glucagon-like Peptide-1 (GLP1) regulates insulin secretion / Vasopressin regulates renal water homeostasis via Aquaporins / neuropeptide binding / Adrenaline,noradrenaline inhibits insulin secretion / ADP signalling through P2Y purinoceptor 12 / G alpha (q) signalling events / G alpha (i) signalling events / Thrombin signalling through proteinase activated receptors (PARs) / Activation of G protein gated Potassium channels / G-protein activation / G beta:gamma signalling through PI3Kgamma / Prostacyclin signalling through prostacyclin receptor / G beta:gamma signalling through PLC beta / ADP signalling through P2Y purinoceptor 1 / Thromboxane signalling through TP receptor / Presynaptic function of Kainate receptors / G beta:gamma signalling through CDC42 / Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits / G alpha (12/13) signalling events / Glucagon-type ligand receptors / G beta:gamma signalling through BTK / eating behavior / ADP signalling through P2Y purinoceptor 12 / Adrenaline,noradrenaline inhibits insulin secretion / Cooperation of PDCL (PhLP1) and TRiC/CCT in G-protein beta folding / Ca2+ pathway / Thrombin signalling through proteinase activated receptors (PARs) / G alpha (z) signalling events / Extra-nuclear estrogen signaling / G alpha (s) signalling events / photoreceptor outer segment membrane / G alpha (q) signalling events / spectrin binding / G alpha (i) signalling events / Glucagon-like Peptide-1 (GLP1) regulates insulin secretion / High laminar flow shear stress activates signaling by PIEZO1 and PECAM1:CDH5:KDR in endothelial cells / Vasopressin regulates renal water homeostasis via Aquaporins / alkylglycerophosphoethanolamine phosphodiesterase activity / regulation of calcium ion transport / photoreceptor outer segment / G protein-coupled receptor signaling pathway, coupled to cyclic nucleotide second messenger / neuropeptide signaling pathway / neuronal dense core vesicle / negative regulation of protein-containing complex assembly / adenylate cyclase inhibitor activity / positive regulation of protein localization to cell cortex / T cell migration / Adenylate cyclase inhibitory pathway / cardiac muscle cell apoptotic process / response to prostaglandin E / D2 dopamine receptor binding / photoreceptor inner segment / G protein-coupled serotonin receptor binding / dendrite membrane / adenylate cyclase regulator activity / adenylate cyclase-inhibiting serotonin receptor signaling pathway / axon terminus / cellular response to forskolin / Peptide ligand-binding receptors / regulation of mitotic spindle organization / adult locomotory behavior / regulation of mitochondrial membrane potential / response to nicotine / Regulation of insulin secretion / defense response / positive regulation of cholesterol biosynthetic process / negative regulation of insulin secretion / G protein-coupled receptor binding / postsynaptic density membrane / adenylate cyclase-inhibiting G protein-coupled receptor signaling pathway / response to peptide hormone / cellular response to growth factor stimulus / adenylate cyclase-modulating G protein-coupled receptor signaling pathway / centriolar satellite / G-protein beta/gamma-subunit complex binding / ADP signalling through P2Y purinoceptor 12 / GDP binding / Adrenaline,noradrenaline inhibits insulin secretion / G alpha (z) signalling events / ADORA2B mediated anti-inflammatory cytokines production Similarity search - Function | ||||||||||||
| Biological species |  Homo sapiens (human) / Phyllomedusa (leaf frogs) / Homo sapiens (human) / Phyllomedusa (leaf frogs) /   | ||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.0 Å | ||||||||||||
 Authors Authors | Wang Y / Zhuang Y / DiBerto JF / Zhou XE / Schmitz GP / Yuan Q / Jain MK / Liu W / Melcher K / Jiang Y ...Wang Y / Zhuang Y / DiBerto JF / Zhou XE / Schmitz GP / Yuan Q / Jain MK / Liu W / Melcher K / Jiang Y / Roth BL / Xu HE | ||||||||||||
| Funding support |  China, 3 items China, 3 items
| ||||||||||||
 Citation Citation |  Journal: Cell / Year: 2023 Journal: Cell / Year: 2023Title: Structures of the entire human opioid receptor family. Authors: Yue Wang / Youwen Zhuang / Jeffrey F DiBerto / X Edward Zhou / Gavin P Schmitz / Qingning Yuan / Manish K Jain / Weiyi Liu / Karsten Melcher / Yi Jiang / Bryan L Roth / H Eric Xu /   Abstract: Opioids are effective analgesics, but their use is beset by serious side effects, including addiction and respiratory depression, which contribute to the ongoing opioid crisis. The human opioid ...Opioids are effective analgesics, but their use is beset by serious side effects, including addiction and respiratory depression, which contribute to the ongoing opioid crisis. The human opioid system contains four opioid receptors (μOR, δOR, κOR, and NOPR) and a set of related endogenous opioid peptides (EOPs), which show distinct selectivity toward their respective opioid receptors (ORs). Despite being key to the development of safer analgesics, the mechanisms of molecular recognition and selectivity of EOPs to ORs remain unclear. Here, we systematically characterize the binding of EOPs to ORs and present five structures of EOP-OR-G complexes, including β-endorphin- and endomorphin-bound μOR, deltorphin-bound δOR, dynorphin-bound κOR, and nociceptin-bound NOPR. These structures, supported by biochemical results, uncover the specific recognition and selectivity of opioid peptides and the conserved mechanism of opioid receptor activation. These results provide a structural framework to facilitate rational design of safer opioid drugs for pain relief. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_28909.map.gz emd_28909.map.gz | 59.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-28909-v30.xml emd-28909-v30.xml emd-28909.xml emd-28909.xml | 24.5 KB 24.5 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_28909.png emd_28909.png | 135 KB | ||
| Filedesc metadata |  emd-28909.cif.gz emd-28909.cif.gz | 7.3 KB | ||
| Others |  emd_28909_half_map_1.map.gz emd_28909_half_map_1.map.gz emd_28909_half_map_2.map.gz emd_28909_half_map_2.map.gz | 49.5 MB 49.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-28909 http://ftp.pdbj.org/pub/emdb/structures/EMD-28909 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-28909 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-28909 | HTTPS FTP |
-Related structure data
| Related structure data |  8f7sMC  8f7qC  8f7rC  8f7wC  8f7xC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_28909.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_28909.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.84 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_28909_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
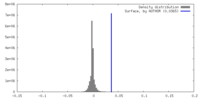
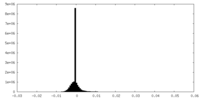
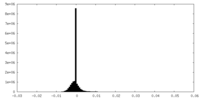
| Density Histograms |
-Half map: #2
| File | emd_28909_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
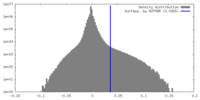
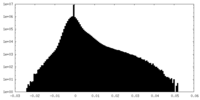
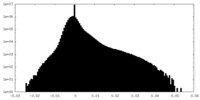
| Density Histograms |
- Sample components
Sample components
-Entire : Gi bound mu-opioid receptor in complex with beta-endorphin
| Entire | Name: Gi bound mu-opioid receptor in complex with beta-endorphin |
|---|---|
| Components |
|
-Supramolecule #1: Gi bound mu-opioid receptor in complex with beta-endorphin
| Supramolecule | Name: Gi bound mu-opioid receptor in complex with beta-endorphin type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#5 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Delta-type opioid receptor
| Macromolecule | Name: Delta-type opioid receptor / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 42.65102 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: DYKDDDDVDA SEPAPSAGAE LQPPLFANAS DAYPSACPSA GANASGPPGA RSASSLALAI AITALYSAVC AVGLLGNVLV MFVIVRYTK MKTATNIYIF SLALAGALAT STLPFQSAKY LMETWPFGEL LCKAVLSIDY YSMFTSIFTL TMMCVDRYIA V CHPVKALD ...String: DYKDDDDVDA SEPAPSAGAE LQPPLFANAS DAYPSACPSA GANASGPPGA RSASSLALAI AITALYSAVC AVGLLGNVLV MFVIVRYTK MKTATNIYIF SLALAGALAT STLPFQSAKY LMETWPFGEL LCKAVLSIDY YSMFTSIFTL TMMCVDRYIA V CHPVKALD FRTPAKAKLI NICIWVLASG VGVPIMVMAV TRPRDGAVVC MLQFPSPSWY WDTVTKICVF LFAFVVPILI IT VCYGLML LRLRSVRLLS GSKEKDRSLR RITRMVLVVV VAFVVCWAPI HIFVIVWTLV DIDRRDPLVV AALHLCIALG YIN SSLNPV LYAFLDENFK RCFRQLCRKP CGRPDPSSFS RAREATARER VTACTPSDGP GGGAAAHHHH HHHH UniProtKB: Delta-type opioid receptor |
-Macromolecule #2: deltorphin
| Macromolecule | Name: deltorphin / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism: Phyllomedusa (leaf frogs) |
| Molecular weight | Theoretical: 781.876 Da |
| Sequence | String: Y(DAL)FEVVG(NH2) UniProtKB: [D-Ala2]-deltorphins |
-Macromolecule #3: Guanine nucleotide-binding protein G(i) subunit alpha-1
| Macromolecule | Name: Guanine nucleotide-binding protein G(i) subunit alpha-1 type: protein_or_peptide / ID: 3 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 40.445059 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGCTLSAEDK AAVERSKMID RNLREDGEKA AREVKLLLLG AGESGKSTIV KQMKIIHEAG YSEEECKQYK AVVYSNTIQS IIAIIRAMG RLKIDFGDSA RADDARQLFV LAGAAEEGFM TAELAGVIKR LWKDSGVQAC FNRSREYQLN DSAAYYLNDL D RIAQPNYI ...String: MGCTLSAEDK AAVERSKMID RNLREDGEKA AREVKLLLLG AGESGKSTIV KQMKIIHEAG YSEEECKQYK AVVYSNTIQS IIAIIRAMG RLKIDFGDSA RADDARQLFV LAGAAEEGFM TAELAGVIKR LWKDSGVQAC FNRSREYQLN DSAAYYLNDL D RIAQPNYI PTQQDVLRTR VKTTGIVETH FTFKDLHFKM FDVGAQRSER KKWIHCFEGV TAIIFCVALS DYDLVLAEDE EM NRMHESM KLFDSICNNK WFTDTSIILF LNKKDLFEEK IKKSPLTICY PEYAGSNTYE EAAAYIQCQF EDLNKRKDTK EIY THFTCS TDTKNVQFVF DAVTDVIIKN NLKDCGLF UniProtKB: Guanine nucleotide-binding protein G(i) subunit alpha-1 |
-Macromolecule #4: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 39.020664 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MHHHHHHHHG SLLQSELDQL RQEAEQLKNQ IRDARKACAD ATLSQITNNI DPVGRIQMRT RRTLRGHLAK IYAMHWGTDS RLLVSASQD GKLIIWDSYT TNKVHAIPLR SSWVMTCAYA PSGNYVACGG LDNICSIYNL KTREGNVRVS RELAGHTGYL S CCRFLDDN ...String: MHHHHHHHHG SLLQSELDQL RQEAEQLKNQ IRDARKACAD ATLSQITNNI DPVGRIQMRT RRTLRGHLAK IYAMHWGTDS RLLVSASQD GKLIIWDSYT TNKVHAIPLR SSWVMTCAYA PSGNYVACGG LDNICSIYNL KTREGNVRVS RELAGHTGYL S CCRFLDDN QIVTSSGDTT CALWDIETGQ QTTTFTGHTG DVMSLSLAPD TRLFVSGACD ASAKLWDVRE GMCRQTFTGH ES DINAICF FPNGNAFATG SDDATCRLFD LRADQELMTY SHDNIICGIT SVSFSKSGRL LLAGYDDFNC NVWDALKADR AGV LAGHDN RVSCLGVTDD GMAVATGSWD SFLKIWN UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 |
-Macromolecule #5: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 7.56375 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MASNNTASIA QARKLVEQLK MEANIDRIKV SKAAADLMAY CEAHAKEDPL LTPVPASENP FREKKFFC UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 |
-Macromolecule #6: CHOLESTEROL
| Macromolecule | Name: CHOLESTEROL / type: ligand / ID: 6 / Number of copies: 5 / Formula: CLR |
|---|---|
| Molecular weight | Theoretical: 386.654 Da |
| Chemical component information |  ChemComp-CLR: |
-Macromolecule #7: PALMITIC ACID
| Macromolecule | Name: PALMITIC ACID / type: ligand / ID: 7 / Number of copies: 12 / Formula: PLM |
|---|---|
| Molecular weight | Theoretical: 256.424 Da |
| Chemical component information |  ChemComp-PLM: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.2 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 23.3 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.8 µm / Nominal defocus min: 0.8 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller
































 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)