[English] 日本語
 Yorodumi
Yorodumi- EMDB-28849: 3-methylcrotonyl-CoA carboxylase in filament, alpha-subunit centered -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | 3-methylcrotonyl-CoA carboxylase in filament, alpha-subunit centered | ||||||||||||||||||||||||
 Map data Map data | mcc | ||||||||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||||||||
 Keywords Keywords | enzyme / multienzyme / multi-enzyme / biotin-dependent / leucine catabolism / PROTEIN FIBRIL / LIGASE | ||||||||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationmethylcrotonoyl-CoA carboxylase activity / mitochondrion / ATP binding / metal ion binding Similarity search - Function | ||||||||||||||||||||||||
| Biological species |  Leishmania tarentolae (eukaryote) Leishmania tarentolae (eukaryote) | ||||||||||||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.9 Å | ||||||||||||||||||||||||
 Authors Authors | Hu JJ / Lee JKJ / Liu YT / Yu C / Huang L / Afasizheva I / Afasizhev R / Zhou ZH | ||||||||||||||||||||||||
| Funding support |  United States, 7 items United States, 7 items
| ||||||||||||||||||||||||
 Citation Citation |  Journal: Structure / Year: 2023 Journal: Structure / Year: 2023Title: Discovery, structure, and function of filamentous 3-methylcrotonyl-CoA carboxylase. Authors: Jason J Hu / Jane K J Lee / Yun-Tao Liu / Clinton Yu / Lan Huang / Inna Aphasizheva / Ruslan Aphasizhev / Z Hong Zhou /  Abstract: 3-methylcrotonyl-CoA carboxylase (MCC) is a biotin-dependent mitochondrial enzyme necessary for leucine catabolism in most organisms. While the crystal structure of recombinant bacterial MCC has been ...3-methylcrotonyl-CoA carboxylase (MCC) is a biotin-dependent mitochondrial enzyme necessary for leucine catabolism in most organisms. While the crystal structure of recombinant bacterial MCC has been characterized, the structure and potential polymerization of native MCC remain elusive. Here, we discovered that native MCC from Leishmania tarentolae (LtMCC) forms filaments, and determined the structures of different filament regions at 3.4, 3.9, and 7.3 Å resolution using cryoEM. αβ LtMCCs assemble in a twisted-stacks architecture, manifesting as supramolecular rods up to 400 nm. Filamentous LtMCCs bind biotin non-covalently and lack coenzyme A. Filaments elongate by stacking αβ LtMCCs onto the exterior α-trimer of the terminal LtMCC. This stacking immobilizes the biotin carboxylase domains, sequestering the enzyme in an inactive state. Our results support a new model for LtMCC catalysis, termed the dual-swinging-domains model, and cast new light on the function of polymerization in the carboxylase superfamily and beyond. | ||||||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_28849.map.gz emd_28849.map.gz | 201.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-28849-v30.xml emd-28849-v30.xml emd-28849.xml emd-28849.xml | 17.9 KB 17.9 KB | Display Display |  EMDB header EMDB header |
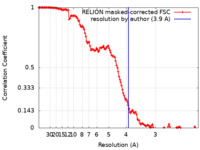
| FSC (resolution estimation) |  emd_28849_fsc.xml emd_28849_fsc.xml | 13.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_28849.png emd_28849.png | 62 KB | ||
| Filedesc metadata |  emd-28849.cif.gz emd-28849.cif.gz | 6.1 KB | ||
| Others |  emd_28849_half_map_1.map.gz emd_28849_half_map_1.map.gz emd_28849_half_map_2.map.gz emd_28849_half_map_2.map.gz | 171.6 MB 171.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-28849 http://ftp.pdbj.org/pub/emdb/structures/EMD-28849 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-28849 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-28849 | HTTPS FTP |
-Related structure data
| Related structure data |  8f41MC  8f3dC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_28849.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_28849.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | mcc | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.1 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: h2
| File | emd_28849_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | h2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
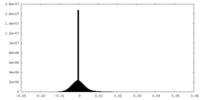
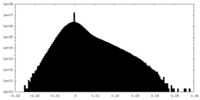
| Density Histograms |
-Half map: h1
| File | emd_28849_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | h1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
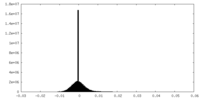
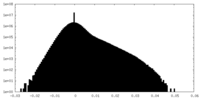
| Density Histograms |
- Sample components
Sample components
-Entire : alpha6beta6 dodecamer in 3-methylcrotonyl-CoA carboxylase filament
| Entire | Name: alpha6beta6 dodecamer in 3-methylcrotonyl-CoA carboxylase filament |
|---|---|
| Components |
|
-Supramolecule #1: alpha6beta6 dodecamer in 3-methylcrotonyl-CoA carboxylase filament
| Supramolecule | Name: alpha6beta6 dodecamer in 3-methylcrotonyl-CoA carboxylase filament type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  Leishmania tarentolae (eukaryote) / Strain: TATII/UC Leishmania tarentolae (eukaryote) / Strain: TATII/UC |
-Macromolecule #1: 3-methylcrotonyl-CoA carboxylase, beta-subunit
| Macromolecule | Name: 3-methylcrotonyl-CoA carboxylase, beta-subunit / type: protein_or_peptide / ID: 1 / Number of copies: 6 / Enantiomer: LEVO / EC number: methylcrotonoyl-CoA carboxylase |
|---|---|
| Source (natural) | Organism:  Leishmania tarentolae (eukaryote) / Strain: TATII/UC Leishmania tarentolae (eukaryote) / Strain: TATII/UC |
| Molecular weight | Theoretical: 61.978711 KDa |
| Sequence | String: YAHHPIDYER STSKSPNILR LPANTSDPTY QENMARMEGL VEQLRARVRY VQAGGVVPEE EAAKAGVSIS SIEADDRVRK LHLSRGKML ARDRIERLID PGTRFLELSQ LAGWDLYWDD KKKEYERCYS GGIVTGIGLV NGVRCMLVAN DATVKGGTYY P ITVKKHLR ...String: YAHHPIDYER STSKSPNILR LPANTSDPTY QENMARMEGL VEQLRARVRY VQAGGVVPEE EAAKAGVSIS SIEADDRVRK LHLSRGKML ARDRIERLID PGTRFLELSQ LAGWDLYWDD KKKEYERCYS GGIVTGIGLV NGVRCMLVAN DATVKGGTYY P ITVKKHLR AQKIAEQNHL PCIYLVDSGG ANLSRQDDVF PDEQHFGRIF YNEAQMSIKS ISQIAVVMGS CTAGGAYVPA MA DENIIVA RNGTIFLGGP PLVLAATGEK VSSEELGGAD VHCRISGVGD HYATDDLHAL YLARRAVANL NLKEHNEARN PTD VKPVPP LYDPRELGGF IPDMLSDVVK SFDVRAIIAR IVDGSRFDEF KALYGNTLVC GFARIEGMQV GIIANQGILY SESA LKGAH FIGLCTQRNV PLLFLQNITG FMVGKKYEEG GIARNGARLV MAVSSAPVPK VTVLIGGSYG AGNYGMCGRA FEPRF LFMW PNARISVMGG TQAATVLTLT NRNLKNASEA EIAAFKDKVK KKYEKEGSCY YSTARLWDDG VIAPEDTRVV VAEALR ATR LAP |
-Macromolecule #2: 3-methylcrotonyl-CoA carboxylase, alpha-subunit
| Macromolecule | Name: 3-methylcrotonyl-CoA carboxylase, alpha-subunit / type: protein_or_peptide / ID: 2 / Number of copies: 6 / Enantiomer: LEVO / EC number: methylcrotonoyl-CoA carboxylase |
|---|---|
| Source (natural) | Organism:  Leishmania tarentolae (eukaryote) / Strain: TATII/UC Leishmania tarentolae (eukaryote) / Strain: TATII/UC |
| Molecular weight | Theoretical: 75.103633 KDa |
| Sequence | String: ERKVEKLLVA NRGEIACRVF RTCREMHIRT VALFCEAERN AKHVAEADEA VCIGPPPAVN SYLRGEHIIS VAKQLNVDAI HPGYGFLSE NASFADAITR SGIEFIGPPA SAISLMGSKS ESKRIMEAAG VPVVPGYYGE NQNVSFLAEE AKKVGFPILI K AVSGGGGK ...String: ERKVEKLLVA NRGEIACRVF RTCREMHIRT VALFCEAERN AKHVAEADEA VCIGPPPAVN SYLRGEHIIS VAKQLNVDAI HPGYGFLSE NASFADAITR SGIEFIGPPA SAISLMGSKS ESKRIMEAAG VPVVPGYYGE NQNVSFLAEE AKKVGFPILI K AVSGGGGK GMKIVERPED FTFMLESAKR EATNFFKDDR VILERYVKRS RHIECQIFFD KHGRGVFFFE RDCSVQRRYQ KV LEEAPAP HLSMETRQRI GEVALQAAKA VGYVGAGTVE FIFDTSTGEF YFMEMNTRLQ VEHPVTEEVC RIKGAPLDLV KLQ IKTAMG KPLTFSQEDV TLVGSCIEAR VYAESPERGF LPESGPLTFI REPFQGVRGP ARTRLDTGFR EGDNVLIHYD PMLA KVISW GRSREEALRG LRQALGEYKV AGINTNIEFL KRCCETPEFA RGGVTTNFIS EHESQLLKSP VVTPEVAAMA ATAWL LNRC DNWRGAFRLN SDTNATVHFY IDDHPVEVRL HTEGANYHKI FFSVWDHDGS FEVCSGPVTS KHRDQKSIVN DFTFLF ENG MHHTVLAVAT EGDVTVIGSF GLHQLRLLPL TDGFGDSSTA GGTSTKIVSP MPGKVSKLLV KSGDLVEKGQ VLVIVEA MK MEHPVRALQD GRVSFLVKEG EVVGGDHVLA TVAEEE UniProtKB: Methylcrotonoyl-coa carboxylase biotinylated subunitprotein-like protein |
-Macromolecule #3: 5-(HEXAHYDRO-2-OXO-1H-THIENO[3,4-D]IMIDAZOL-6-YL)PENTANAL
| Macromolecule | Name: 5-(HEXAHYDRO-2-OXO-1H-THIENO[3,4-D]IMIDAZOL-6-YL)PENTANAL type: ligand / ID: 3 / Number of copies: 6 / Formula: BTI |
|---|---|
| Molecular weight | Theoretical: 228.311 Da |
| Chemical component information |  ChemComp-BTI: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | filament |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277.15 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: AB INITIO MODEL |
|---|---|
| Output model |  PDB-8f41: |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)