[English] 日本語
 Yorodumi
Yorodumi- EMDB-28846: 3-methylcrotonyl-CoA carboxylase in filament, beta-subunit centered -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | 3-methylcrotonyl-CoA carboxylase in filament, beta-subunit centered | ||||||||||||||||||||||||
 Map data Map data | beta-subunit centered map | ||||||||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||||||||
 Keywords Keywords | enzyme / multienzyme / multi-enzyme / biotin-dependent / leucine catabolism / PROTEIN FIBRIL / LIGASE | ||||||||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationmethylcrotonoyl-CoA carboxylase activity / mitochondrion / ATP binding / metal ion binding Similarity search - Function | ||||||||||||||||||||||||
| Biological species |  Leishmania tarentolae (eukaryote) Leishmania tarentolae (eukaryote) | ||||||||||||||||||||||||
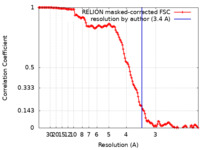
| Method | single particle reconstruction / cryo EM / Resolution: 3.4 Å | ||||||||||||||||||||||||
 Authors Authors | Hu JJ / Lee JKJ / Liu YT / Yu C / Huang L / Afasizheva I / Afasizhev R / Zhou ZH | ||||||||||||||||||||||||
| Funding support |  United States, 7 items United States, 7 items
| ||||||||||||||||||||||||
 Citation Citation |  Journal: Structure / Year: 2023 Journal: Structure / Year: 2023Title: Discovery, structure, and function of filamentous 3-methylcrotonyl-CoA carboxylase. Authors: Jason J Hu / Jane K J Lee / Yun-Tao Liu / Clinton Yu / Lan Huang / Inna Aphasizheva / Ruslan Aphasizhev / Z Hong Zhou /  Abstract: 3-methylcrotonyl-CoA carboxylase (MCC) is a biotin-dependent mitochondrial enzyme necessary for leucine catabolism in most organisms. While the crystal structure of recombinant bacterial MCC has been ...3-methylcrotonyl-CoA carboxylase (MCC) is a biotin-dependent mitochondrial enzyme necessary for leucine catabolism in most organisms. While the crystal structure of recombinant bacterial MCC has been characterized, the structure and potential polymerization of native MCC remain elusive. Here, we discovered that native MCC from Leishmania tarentolae (LtMCC) forms filaments, and determined the structures of different filament regions at 3.4, 3.9, and 7.3 Å resolution using cryoEM. αβ LtMCCs assemble in a twisted-stacks architecture, manifesting as supramolecular rods up to 400 nm. Filamentous LtMCCs bind biotin non-covalently and lack coenzyme A. Filaments elongate by stacking αβ LtMCCs onto the exterior α-trimer of the terminal LtMCC. This stacking immobilizes the biotin carboxylase domains, sequestering the enzyme in an inactive state. Our results support a new model for LtMCC catalysis, termed the dual-swinging-domains model, and cast new light on the function of polymerization in the carboxylase superfamily and beyond. #1:  Journal: To Be Published Journal: To Be PublishedTitle: Discovery, Structure, and Function of Filamentous 3-Methylcrotonyl-CoA Carboxylase Authors: Hu JJ / Lee JKJ / Liu YT / Yu C / Huang L / Afasizheva I / Afasizhev R / Zhou ZH | ||||||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_28846.map.gz emd_28846.map.gz | 202 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-28846-v30.xml emd-28846-v30.xml emd-28846.xml emd-28846.xml | 19.2 KB 19.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_28846_fsc.xml emd_28846_fsc.xml | 13.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_28846.png emd_28846.png | 71.1 KB | ||
| Filedesc metadata |  emd-28846.cif.gz emd-28846.cif.gz | 6.4 KB | ||
| Others |  emd_28846_half_map_1.map.gz emd_28846_half_map_1.map.gz emd_28846_half_map_2.map.gz emd_28846_half_map_2.map.gz | 171.6 MB 171.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-28846 http://ftp.pdbj.org/pub/emdb/structures/EMD-28846 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-28846 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-28846 | HTTPS FTP |
-Related structure data
| Related structure data |  8f3dMC  8f41C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_28846.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_28846.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | beta-subunit centered map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.1 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: half 1 of beta-subunit centered map
| File | emd_28846_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half 1 of beta-subunit centered map | ||||||||||||
| Projections & Slices |
| ||||||||||||
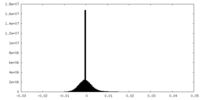
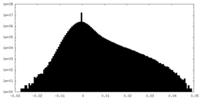
| Density Histograms |
-Half map: half 2 of beta-subunit centered map
| File | emd_28846_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half 2 of beta-subunit centered map | ||||||||||||
| Projections & Slices |
| ||||||||||||
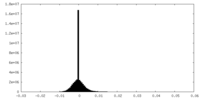
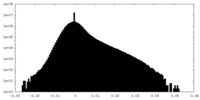
| Density Histograms |
- Sample components
Sample components
-Entire : alpha6beta6 dodecamer in 3-methylcrotonyl-CoA filament
| Entire | Name: alpha6beta6 dodecamer in 3-methylcrotonyl-CoA filament |
|---|---|
| Components |
|
-Supramolecule #1: alpha6beta6 dodecamer in 3-methylcrotonyl-CoA filament
| Supramolecule | Name: alpha6beta6 dodecamer in 3-methylcrotonyl-CoA filament type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  Leishmania tarentolae (eukaryote) / Strain: TATII/UC Leishmania tarentolae (eukaryote) / Strain: TATII/UC |
-Macromolecule #1: 3-methylcrotonyl-CoA carboxylase beta-subunit
| Macromolecule | Name: 3-methylcrotonyl-CoA carboxylase beta-subunit / type: protein_or_peptide / ID: 1 / Number of copies: 6 / Enantiomer: LEVO / EC number: methylcrotonoyl-CoA carboxylase |
|---|---|
| Source (natural) | Organism:  Leishmania tarentolae (eukaryote) / Strain: TATII/UC Leishmania tarentolae (eukaryote) / Strain: TATII/UC |
| Molecular weight | Theoretical: 76.468469 KDa |
| Sequence | String: MDKTHVLRNT TRGMFRTSMM RVLGRPGSLS VTASAGGTAC VPMAALLRRS ATPAAASATA ASAMSALWCT RCFESSSAGS PPKVSDAGM KERSTQEPCV AATPTPSDTA ATLAAAATPA PAAPIKSKGV DYARLYAHHP IDYERSTSKS PNILRLPANT S DPTYQENM ...String: MDKTHVLRNT TRGMFRTSMM RVLGRPGSLS VTASAGGTAC VPMAALLRRS ATPAAASATA ASAMSALWCT RCFESSSAGS PPKVSDAGM KERSTQEPCV AATPTPSDTA ATLAAAATPA PAAPIKSKGV DYARLYAHHP IDYERSTSKS PNILRLPANT S DPTYQENM ARMEGLVEQL RARVRYVQAG GVVPEEEAAK AGVSISSIEA DDRVRKLHLS RGKMLARDRI ERLIDPGTRF LE LSQLAGW DLYWDDKKKE YERCYSGGIV TGIGLVNGVR CMLVANDATV KGGTYYPITV KKHLRAQKIA EQNHLPCIYL VDS GGANLS RQDDVFPDEQ HFGRIFYNEA QMSIKSISQI AVVMGSCTAG GAYVPAMADE NIIVARNGTI FLGGPPLVLA ATGE KVSSE ELGGADVHCR ISGVGDHYAT DDLHALYLAR RAVANLNLKE HNEARNPTDV KPVPPLYDPR ELGGFIPDML SDVVK SFDV RAIIARIVDG SRFDEFKALY GNTLVCGFAR IEGMQVGIIA NQGILYSESA LKGAHFIGLC TQRNVPLLFL QNITGF MVG KKYEEGGIAR NGARLVMAVS SAPVPKVTVL IGGSYGAGNY GMCGRAFEPR FLFMWPNARI SVMGGTQAAT VLTLTNR NL KNASEAEIAA FKDKVKKKYE KEGSCYYSTA RLWDDGVIAP EDTRVVVAEA LRATRLAPME KRERV |
-Macromolecule #2: 3-methylcrotonyl-CoA carboxylase alpha-subunit
| Macromolecule | Name: 3-methylcrotonyl-CoA carboxylase alpha-subunit / type: protein_or_peptide / ID: 2 / Number of copies: 6 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Leishmania tarentolae (eukaryote) / Strain: TATII/UC Leishmania tarentolae (eukaryote) / Strain: TATII/UC |
| Molecular weight | Theoretical: 76.28307 KDa |
| Sequence | String: MLRYTGLWRE RKVEKLLVAN RGEIACRVFR TCREMHIRTV ALFCEAERNA KHVAEADEAV CIGPPPAVNS YLRGEHIISV AKQLNVDAI HPGYGFLSEN ASFADAITRS GIEFIGPPAS AISLMGSKSE SKRIMEAAGV PVVPGYYGEN QNVSFLAEEA K KVGFPILI ...String: MLRYTGLWRE RKVEKLLVAN RGEIACRVFR TCREMHIRTV ALFCEAERNA KHVAEADEAV CIGPPPAVNS YLRGEHIISV AKQLNVDAI HPGYGFLSEN ASFADAITRS GIEFIGPPAS AISLMGSKSE SKRIMEAAGV PVVPGYYGEN QNVSFLAEEA K KVGFPILI KAVSGGGGKG MKIVERPEDF TFMLESAKRE ATNFFKDDRV ILERYVKRSR HIECQIFFDK HGRGVFFFER DC SVQRRYQ KVLEEAPAPH LSMETRQRIG EVALQAAKAV GYVGAGTVEF IFDTSTGEFY FMEMNTRLQV EHPVTEEVCR IKG APLDLV KLQIKTAMGK PLTFSQEDVT LVGSCIEARV YAESPERGFL PESGPLTFIR EPFQGVRGPA RTRLDTGFRE GDNV LIHYD PMLAKVISWG RSREEALRGL RQALGEYKVA GINTNIEFLK RCCETPEFAR GGVTTNFISE HESQLLKSPV VTPEV AAMA ATAWLLNRCD NWRGAFRLNS DTNATVHFYI DDHPVEVRLH TEGANYHKIF FSVWDHDGSF EVCSGPVTSK HRDQKS IVN DFTFLFENGM HHTVLAVATE GDVTVIGSFG LHQLRLLPLT DGFGDSSTAG GTSTKIVSPM PGKVSKLLVK SGDLVEK GQ VLVIVEAMKM EHPVRALQDG RVSFLVKEGE VVGGDHVLAT VAEEE UniProtKB: Methylcrotonoyl-coa carboxylase biotinylated subunitprotein-like protein |
-Macromolecule #3: 5-(HEXAHYDRO-2-OXO-1H-THIENO[3,4-D]IMIDAZOL-6-YL)PENTANAL
| Macromolecule | Name: 5-(HEXAHYDRO-2-OXO-1H-THIENO[3,4-D]IMIDAZOL-6-YL)PENTANAL type: ligand / ID: 3 / Number of copies: 6 / Formula: BTI |
|---|---|
| Molecular weight | Theoretical: 228.311 Da |
| Chemical component information |  ChemComp-BTI: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | filament |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277.15 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: AB INITIO MODEL |
|---|---|
| Output model |  PDB-8f3d: |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)