[English] 日本語
 Yorodumi
Yorodumi- EMDB-28273: CryoEM structure of the high pH nitrogenase MoFe-protein under no... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | CryoEM structure of the high pH nitrogenase MoFe-protein under non-turnover conditions | ||||||||||||
 Map data Map data | |||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | nitrogenase / nitrogen fixation / reductase / MoFe / OXIDOREDUCTASE | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationmolybdenum-iron nitrogenase complex / nitrogenase / nitrogenase activity / nitrogen fixation / iron-sulfur cluster binding / ATP binding / metal ion binding Similarity search - Function | ||||||||||||
| Biological species |  Azotobacter vinelandii (bacteria) Azotobacter vinelandii (bacteria) | ||||||||||||
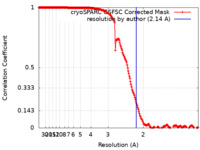
| Method | single particle reconstruction / cryo EM / Resolution: 2.14 Å | ||||||||||||
 Authors Authors | Warmack RA / Maggiolo AO / Rees DC | ||||||||||||
| Funding support |  United States, 3 items United States, 3 items
| ||||||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2023 Journal: Nat Commun / Year: 2023Title: Structural consequences of turnover-induced homocitrate loss in nitrogenase. Authors: Rebeccah A Warmack / Ailiena O Maggiolo / Andres Orta / Belinda B Wenke / James B Howard / Douglas C Rees /  Abstract: Nitrogenase catalyzes the ATP-dependent reduction of dinitrogen to ammonia during the process of biological nitrogen fixation that is essential for sustaining life. The active site FeMo-cofactor ...Nitrogenase catalyzes the ATP-dependent reduction of dinitrogen to ammonia during the process of biological nitrogen fixation that is essential for sustaining life. The active site FeMo-cofactor contains a [7Fe:1Mo:9S:1C] metallocluster coordinated with an R-homocitrate (HCA) molecule. Here, we establish through single particle cryoEM and chemical analysis of two forms of the Azotobacter vinelandii MoFe-protein - a high pH turnover inactivated species and a ∆NifV variant that cannot synthesize HCA - that loss of HCA is coupled to α-subunit domain and FeMo-cofactor disordering, and formation of a histidine coordination site. We further find a population of the ∆NifV variant complexed to an endogenous protein identified through structural and proteomic approaches as the uncharacterized protein NafT. Recognition by endogenous NafT demonstrates the physiological relevance of the HCA-compromised form, perhaps for cofactor insertion or repair. Our results point towards a dynamic active site in which HCA plays a role in enabling nitrogenase catalysis by facilitating activation of the FeMo-cofactor from a relatively stable form to a state capable of reducing dinitrogen under ambient conditions. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_28273.map.gz emd_28273.map.gz | 374.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-28273-v30.xml emd-28273-v30.xml emd-28273.xml emd-28273.xml | 20.3 KB 20.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_28273_fsc.xml emd_28273_fsc.xml | 18.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_28273.png emd_28273.png | 87 KB | ||
| Filedesc metadata |  emd-28273.cif.gz emd-28273.cif.gz | 6.6 KB | ||
| Others |  emd_28273_half_map_1.map.gz emd_28273_half_map_1.map.gz emd_28273_half_map_2.map.gz emd_28273_half_map_2.map.gz | 676.4 MB 676.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-28273 http://ftp.pdbj.org/pub/emdb/structures/EMD-28273 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-28273 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-28273 | HTTPS FTP |
-Related structure data
| Related structure data |  8enmMC  8crsC  8dbxC  8enlC  8ennC  8enoC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_28273.map.gz / Format: CCP4 / Size: 729 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_28273.map.gz / Format: CCP4 / Size: 729 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.65 Å | ||||||||||||||||||||||||||||||||||||
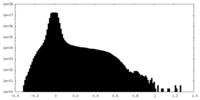
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_28273_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
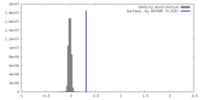
| Density Histograms |
-Half map: #1
| File | emd_28273_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
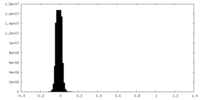
| Density Histograms |
- Sample components
Sample components
-Entire : Heterotetrameric nitrogenase MoFe-protein
| Entire | Name: Heterotetrameric nitrogenase MoFe-protein |
|---|---|
| Components |
|
-Supramolecule #1: Heterotetrameric nitrogenase MoFe-protein
| Supramolecule | Name: Heterotetrameric nitrogenase MoFe-protein / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  Azotobacter vinelandii (bacteria) Azotobacter vinelandii (bacteria) |
-Macromolecule #1: Nitrogenase molybdenum-iron protein alpha chain
| Macromolecule | Name: Nitrogenase molybdenum-iron protein alpha chain / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO / EC number: nitrogenase |
|---|---|
| Source (natural) | Organism:  Azotobacter vinelandii (bacteria) Azotobacter vinelandii (bacteria) |
| Molecular weight | Theoretical: 55.363043 KDa |
| Sequence | String: MTGMSREEVE SLIQEVLEVY PEKARKDRNK HLAVNDPAVT QSKKCIISNK KSQPGLMTIR GCAYAGSKGV VWGPIKDMIH ISHGPVGCG QYSRAGRRNY YIGTTGVNAF VTMNFTSDFQ EKDIVFGGDK KLAKLIDEVE TLFPLNKGIS VQSECPIGLI G DDIESVSK ...String: MTGMSREEVE SLIQEVLEVY PEKARKDRNK HLAVNDPAVT QSKKCIISNK KSQPGLMTIR GCAYAGSKGV VWGPIKDMIH ISHGPVGCG QYSRAGRRNY YIGTTGVNAF VTMNFTSDFQ EKDIVFGGDK KLAKLIDEVE TLFPLNKGIS VQSECPIGLI G DDIESVSK VKGAELSKTI VPVRCEGFRG VSQSLGHHIA NDAVRDWVLG KRDEDTTFAS TPYDVAIIGD YNIGGDAWSS RI LLEEMGL RCVAQWSGDG SISEIELTPK VKLNLVHCYR SMNYISRHME EKYGIPWMEY NFFGPTKTIE SLRAIAAKFD ESI QKKCEE VIAKYKPEWE AVVAKYRPRL EGKRVMLYIG GLRPRHVIGA YEDLGMEVVG TGYEFAHNDD YDRTMKEMGD STLL YDDVT GYEFEEFVKR IKPDLIGSGI KEKFIFQKMG IPFREMHSWD YSGPYHGFDG FAIFARDMDM TLNNPCWKKL QAPWE ASEG AEKVAASA UniProtKB: Nitrogenase molybdenum-iron protein alpha chain |
-Macromolecule #2: Nitrogenase molybdenum-iron protein beta chain
| Macromolecule | Name: Nitrogenase molybdenum-iron protein beta chain / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO / EC number: nitrogenase |
|---|---|
| Source (natural) | Organism:  Azotobacter vinelandii (bacteria) Azotobacter vinelandii (bacteria) |
| Molecular weight | Theoretical: 59.535879 KDa |
| Sequence | String: MSQQVDKIKA SYPLFLDQDY KDMLAKKRDG FEEKYPQDKI DEVFQWTTTK EYQELNFQRE ALTVNPAKAC QPLGAVLCAL GFEKTMPYV HGSQGCVAYF RSYFNRHFRE PVSCVSDSMT EDAAVFGGQQ NMKDGLQNCK ATYKPDMIAV STTCMAEVIG D DLNAFINN ...String: MSQQVDKIKA SYPLFLDQDY KDMLAKKRDG FEEKYPQDKI DEVFQWTTTK EYQELNFQRE ALTVNPAKAC QPLGAVLCAL GFEKTMPYV HGSQGCVAYF RSYFNRHFRE PVSCVSDSMT EDAAVFGGQQ NMKDGLQNCK ATYKPDMIAV STTCMAEVIG D DLNAFINN SKKEGFIPDE FPVPFAHTPS FVGSHVTGWD NMFEGIARYF TLKSMDDKVV GSNKKINIVP GFETYLGNFR VI KRMLSEM GVGYSLLSDP EEVLDTPADG QFRMYAGGTT QEEMKDAPNA LNTVLLQPWH LEKTKKFVEG TWKHEVPKLN IPM GLDWTD EFLMKVSEIS GQPIPASLTK ERGRLVDMMT DSHTWLHGKR FALWGDPDFV MGLVKFLLEL GCEPVHILCH NGNK RWKKA VDAILAASPY GKNATVYIGK DLWHLRSLVF TDKPDFMIGN SYGKFIQRDT LHKGKEFEVP LIRIGFPIFD RHHLH RSTT LGYEGAMQIL TTLVNSILER LDEETRGMQA TDYNHDLVR UniProtKB: Nitrogenase molybdenum-iron protein beta chain |
-Macromolecule #3: iron-sulfur-molybdenum cluster with interstitial carbon
| Macromolecule | Name: iron-sulfur-molybdenum cluster with interstitial carbon type: ligand / ID: 3 / Number of copies: 2 / Formula: ICS |
|---|---|
| Molecular weight | Theoretical: 787.451 Da |
| Chemical component information | 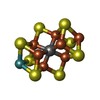 ChemComp-ICE: |
-Macromolecule #4: 3-HYDROXY-3-CARBOXY-ADIPIC ACID
| Macromolecule | Name: 3-HYDROXY-3-CARBOXY-ADIPIC ACID / type: ligand / ID: 4 / Number of copies: 2 / Formula: HCA |
|---|---|
| Molecular weight | Theoretical: 206.15 Da |
| Chemical component information | 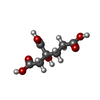 ChemComp-HCA: |
-Macromolecule #5: FE(8)-S(7) CLUSTER
| Macromolecule | Name: FE(8)-S(7) CLUSTER / type: ligand / ID: 5 / Number of copies: 2 / Formula: CLF |
|---|---|
| Molecular weight | Theoretical: 671.215 Da |
| Chemical component information |  ChemComp-CLF: |
-Macromolecule #6: FE (III) ION
| Macromolecule | Name: FE (III) ION / type: ligand / ID: 6 / Number of copies: 2 / Formula: FE |
|---|---|
| Molecular weight | Theoretical: 55.845 Da |
-Macromolecule #7: water
| Macromolecule | Name: water / type: ligand / ID: 7 / Number of copies: 1693 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 4.75 mg/mL |
|---|---|
| Buffer | pH: 7.8 |
| Vitrification | Cryogen name: ETHANE-PROPANE |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: BRIGHT FIELD / Nominal defocus max: -3.0 µm / Nominal defocus min: -0.8 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)