[English] 日本語
 Yorodumi
Yorodumi- EMDB-28101: Structure of single homo-hexameric Holliday junction ATP-dependen... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of single homo-hexameric Holliday junction ATP-dependent DNA helicase RuvB motor | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Holliday Junction / AAA+ ATPase / RuvB / Homo-hexamer / DNA recombination / STRUCTURAL PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationHolliday junction resolvase complex / four-way junction helicase activity / four-way junction DNA binding / Hydrolases; Acting on acid anhydrides; Acting on acid anhydrides to facilitate cellular and subcellular movement / DNA recombination / DNA repair / ATP hydrolysis activity / ATP binding / cytoplasm Similarity search - Function | |||||||||
| Biological species |   Thermus thermophilus HB8 (bacteria) Thermus thermophilus HB8 (bacteria) | |||||||||
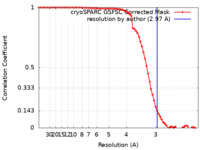
| Method | single particle reconstruction / cryo EM / Resolution: 2.97 Å | |||||||||
 Authors Authors | Shen ZF / Rish AD / Fu TM | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2023 Journal: Nat Commun / Year: 2023Title: Molecular mechanisms of Holliday junction branch migration catalyzed by an asymmetric RuvB hexamer. Authors: Anthony D Rish / Zhangfei Shen / Zhenhang Chen / Nan Zhang / Qingfei Zheng / Tian-Min Fu /  Abstract: The Holliday junction (HJ) is a DNA intermediate of homologous recombination, involved in many fundamental physiological processes. RuvB, an ATPase motor protein, drives branch migration of the ...The Holliday junction (HJ) is a DNA intermediate of homologous recombination, involved in many fundamental physiological processes. RuvB, an ATPase motor protein, drives branch migration of the Holliday junction with a mechanism that had yet to be elucidated. Here we report two cryo-EM structures of RuvB, providing a comprehensive understanding of HJ branch migration. RuvB assembles into a spiral staircase, ring-like hexamer, encircling dsDNA. Four protomers of RuvB contact the DNA backbone with a translocation step size of 2 nucleotides. The variation of nucleotide-binding states in RuvB supports a sequential model for ATP hydrolysis and nucleotide recycling, which occur at separate, singular positions. RuvB's asymmetric assembly also explains the 6:4 stoichiometry between the RuvB/RuvA complex, which coordinates HJ migration in bacteria. Taken together, we provide a mechanistic understanding of HJ branch migration facilitated by RuvB, which may be universally shared by prokaryotic and eukaryotic organisms. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_28101.map.gz emd_28101.map.gz | 36.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-28101-v30.xml emd-28101-v30.xml emd-28101.xml emd-28101.xml | 20.3 KB 20.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_28101_fsc.xml emd_28101_fsc.xml | 7.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_28101.png emd_28101.png | 66.3 KB | ||
| Filedesc metadata |  emd-28101.cif.gz emd-28101.cif.gz | 6.4 KB | ||
| Others |  emd_28101_half_map_1.map.gz emd_28101_half_map_1.map.gz emd_28101_half_map_2.map.gz emd_28101_half_map_2.map.gz | 35.7 MB 35.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-28101 http://ftp.pdbj.org/pub/emdb/structures/EMD-28101 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-28101 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-28101 | HTTPS FTP |
-Related structure data
| Related structure data |  8efvMC  8efyC  8gh8C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_28101.map.gz / Format: CCP4 / Size: 38.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_28101.map.gz / Format: CCP4 / Size: 38.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.08 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_28101_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_28101_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Single homo-hexameric AAA+ ATPase RuvB motor
| Entire | Name: Single homo-hexameric AAA+ ATPase RuvB motor |
|---|---|
| Components |
|
-Supramolecule #1: Single homo-hexameric AAA+ ATPase RuvB motor
| Supramolecule | Name: Single homo-hexameric AAA+ ATPase RuvB motor / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 |
|---|---|
| Source (natural) | Organism:   Thermus thermophilus HB8 (bacteria) Thermus thermophilus HB8 (bacteria) |
-Macromolecule #1: Holliday junction ATP-dependent DNA helicase RuvB
| Macromolecule | Name: Holliday junction ATP-dependent DNA helicase RuvB / type: protein_or_peptide / ID: 1 / Number of copies: 6 / Enantiomer: LEVO / EC number: DNA helicase |
|---|---|
| Source (natural) | Organism:   Thermus thermophilus HB8 (bacteria) / Strain: ATCC 27634 / DSM 579 / HB8 Thermus thermophilus HB8 (bacteria) / Strain: ATCC 27634 / DSM 579 / HB8 |
| Molecular weight | Theoretical: 36.024688 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MEDLALRPKT LDEYIGQERL KQKLRVYLEA AKARKEPLEH LLLFGPPGLG KTTLAHVIAH ELGVNLRVTS GPAIEKPGDL AAILANSLE EGDILFIDEI HRLSRQAEEH LYPAMEDFVM DIVIGQGPAA RTIRLELPRF TLIGATTRPG LITAPLLSRF G IVEHLEYY ...String: MEDLALRPKT LDEYIGQERL KQKLRVYLEA AKARKEPLEH LLLFGPPGLG KTTLAHVIAH ELGVNLRVTS GPAIEKPGDL AAILANSLE EGDILFIDEI HRLSRQAEEH LYPAMEDFVM DIVIGQGPAA RTIRLELPRF TLIGATTRPG LITAPLLSRF G IVEHLEYY TPEELAQGVM RDARLLGVRI TEEAALEIGR RSRGTMRVAK RLFRRVRDFA QVAGEEVITR ERALEALAAL GL DELGLEK RDREILEVLI LRFGGGPVGL ATLATALSED PGTLEEVHEP YLIRQGLLKR TPRGRVATEL AYRHLGYPPP VGP LLEP UniProtKB: Holliday junction branch migration complex subunit RuvB |
-Macromolecule #2: 49-mer DNA
| Macromolecule | Name: 49-mer DNA / type: dna / ID: 2 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism:   Thermus thermophilus HB8 (bacteria) Thermus thermophilus HB8 (bacteria) |
| Molecular weight | Theoretical: 15.146732 KDa |
| Sequence | String: (DA)(DG)(DA)(DA)(DT)(DC)(DT)(DG)(DC)(DC) (DG)(DA)(DG)(DA)(DG)(DA)(DC)(DC)(DG)(DA) (DG)(DC)(DA)(DG)(DA)(DA)(DT)(DT)(DC) (DT)(DA)(DT)(DG)(DT)(DG)(DT)(DT)(DT)(DA) (DC) (DC)(DA)(DA)(DG)(DC)(DG)(DC)(DT) (DG) |
-Macromolecule #3: 51-mer DNA
| Macromolecule | Name: 51-mer DNA / type: dna / ID: 3 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism:   Thermus thermophilus HB8 (bacteria) Thermus thermophilus HB8 (bacteria) |
| Molecular weight | Theoretical: 15.69107 KDa |
| Sequence | String: (DC)(DA)(DG)(DC)(DG)(DC)(DT)(DT)(DG)(DG) (DT)(DA)(DA)(DA)(DC)(DA)(DC)(DA)(DT)(DA) (DG)(DA)(DA)(DT)(DT)(DC)(DT)(DG)(DC) (DT)(DC)(DT)(DC)(DG)(DG)(DT)(DC)(DT)(DG) (DA) (DG)(DC)(DC)(DG)(DT)(DC)(DT)(DA) (DA)(DG)(DA) |
-Macromolecule #4: PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER
| Macromolecule | Name: PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER / type: ligand / ID: 4 / Number of copies: 3 / Formula: AGS |
|---|---|
| Molecular weight | Theoretical: 523.247 Da |
| Chemical component information |  ChemComp-AGS: |
-Macromolecule #5: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 5 / Number of copies: 4 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Macromolecule #6: ADENOSINE-5'-DIPHOSPHATE
| Macromolecule | Name: ADENOSINE-5'-DIPHOSPHATE / type: ligand / ID: 6 / Number of copies: 3 / Formula: ADP |
|---|---|
| Molecular weight | Theoretical: 427.201 Da |
| Chemical component information |  ChemComp-ADP: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)