[English] 日本語
 Yorodumi
Yorodumi- EMDB-28074: CryoEM of the soluble OPA1 dimer from the apo helical assembly on... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | CryoEM of the soluble OPA1 dimer from the apo helical assembly on a lipid membrane | |||||||||
 Map data Map data | sOPA1 apo helical assembly full map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | GTPase / Dynamin-family protein / mitochondrial fusion protein / mitochondria / Optic Atrophy / LIPID BINDING PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationRegulation of Apoptosis / mitochondrial inner membrane fusion / membrane tubulation / membrane bending activity / inner mitochondrial membrane organization / GTPase-dependent fusogenic activity / dynamin GTPase / cristae formation / peroxisome fission / : ...Regulation of Apoptosis / mitochondrial inner membrane fusion / membrane tubulation / membrane bending activity / inner mitochondrial membrane organization / GTPase-dependent fusogenic activity / dynamin GTPase / cristae formation / peroxisome fission / : / phosphatidic acid binding / cardiolipin binding / mitochondrial fission / GTP metabolic process / mitochondrial fusion / axonal transport of mitochondrion / negative regulation of release of cytochrome c from mitochondria / mitochondrial crista / intracellular distribution of mitochondria / positive regulation of interleukin-17 production / protein complex oligomerization / positive regulation of T-helper 17 cell lineage commitment / negative regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway / visual perception / axon cytoplasm / Mitochondrial protein degradation / mitochondrion organization / neural tube closure / mitochondrial membrane / mitochondrial intermembrane space / endocytosis / cellular senescence / microtubule binding / mitochondrial outer membrane / microtubule / mitochondrial inner membrane / GTPase activity / apoptotic process / dendrite / negative regulation of apoptotic process / GTP binding / magnesium ion binding / mitochondrion / nucleoplasm / membrane / cytosol / cytoplasm Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
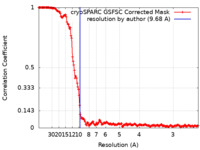
| Method | helical reconstruction / cryo EM / Resolution: 9.68 Å | |||||||||
 Authors Authors | Nyenhuis SB / Wu X / Stanton AE / Strub MP / Yim YI / Canagarajah B / Hinshaw JE | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Nature / Year: 2023 Journal: Nature / Year: 2023Title: OPA1 helical structures give perspective to mitochondrial dysfunction. Authors: Sarah B Nyenhuis / Xufeng Wu / Marie-Paule Strub / Yang-In Yim / Abigail E Stanton / Valentina Baena / Zulfeqhar A Syed / Bertram Canagarajah / John A Hammer / Jenny E Hinshaw /  Abstract: Dominant optic atrophy is one of the leading causes of childhood blindness. Around 60-80% of cases are caused by mutations of the gene that encodes optic atrophy protein 1 (OPA1), a protein that has ...Dominant optic atrophy is one of the leading causes of childhood blindness. Around 60-80% of cases are caused by mutations of the gene that encodes optic atrophy protein 1 (OPA1), a protein that has a key role in inner mitochondrial membrane fusion and remodelling of cristae and is crucial for the dynamic organization and regulation of mitochondria. Mutations in OPA1 result in the dysregulation of the GTPase-mediated fusion process of the mitochondrial inner and outer membranes. Here we used cryo-electron microscopy methods to solve helical structures of OPA1 assembled on lipid membrane tubes, in the presence and absence of nucleotide. These helical assemblies organize into densely packed protein rungs with minimal inter-rung connectivity, and exhibit nucleotide-dependent dimerization of the GTPase domains-a hallmark of the dynamin superfamily of proteins. OPA1 also contains several unique secondary structures in the paddle domain that strengthen its membrane association, including membrane-inserting helices. The structural features identified in this study shed light on the effects of pathogenic point mutations on protein folding, inter-protein assembly and membrane interactions. Furthermore, mutations that disrupt the assembly interfaces and membrane binding of OPA1 cause mitochondrial fragmentation in cell-based assays, providing evidence of the biological relevance of these interactions. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_28074.map.gz emd_28074.map.gz | 204.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-28074-v30.xml emd-28074-v30.xml emd-28074.xml emd-28074.xml | 15.3 KB 15.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_28074_fsc.xml emd_28074_fsc.xml | 15.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_28074.png emd_28074.png | 90.4 KB | ||
| Masks |  emd_28074_msk_1.map emd_28074_msk_1.map | 421.9 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-28074.cif.gz emd-28074.cif.gz | 5.9 KB | ||
| Others |  emd_28074_half_map_1.map.gz emd_28074_half_map_1.map.gz emd_28074_half_map_2.map.gz emd_28074_half_map_2.map.gz | 391.1 MB 391.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-28074 http://ftp.pdbj.org/pub/emdb/structures/EMD-28074 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-28074 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-28074 | HTTPS FTP |
-Validation report
| Summary document |  emd_28074_validation.pdf.gz emd_28074_validation.pdf.gz | 1.3 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_28074_full_validation.pdf.gz emd_28074_full_validation.pdf.gz | 1.3 MB | Display | |
| Data in XML |  emd_28074_validation.xml.gz emd_28074_validation.xml.gz | 24.4 KB | Display | |
| Data in CIF |  emd_28074_validation.cif.gz emd_28074_validation.cif.gz | 31.8 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-28074 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-28074 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-28074 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-28074 | HTTPS FTP |
-Related structure data
| Related structure data |  8ef7MC  8efsMC  8eftMC  8eewC  8effC  8efrC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_28074.map.gz / Format: CCP4 / Size: 421.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_28074.map.gz / Format: CCP4 / Size: 421.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | sOPA1 apo helical assembly full map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.2518 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_28074_msk_1.map emd_28074_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: sOPA1 apo helical assembly half map A
| File | emd_28074_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | sOPA1 apo helical assembly half map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: sOPA1 apo helical assembly half map B
| File | emd_28074_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | sOPA1 apo helical assembly half map B | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : cryoEM of the soluble OPA1 helical assembly on a lipid membrane i...
| Entire | Name: cryoEM of the soluble OPA1 helical assembly on a lipid membrane in the apo state |
|---|---|
| Components |
|
-Supramolecule #1: cryoEM of the soluble OPA1 helical assembly on a lipid membrane i...
| Supramolecule | Name: cryoEM of the soluble OPA1 helical assembly on a lipid membrane in the apo state type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Dynamin-like 120 kDa protein, form S1
| Macromolecule | Name: Dynamin-like 120 kDa protein, form S1 / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 89.262516 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: ATDRGSESDK HFRKVSDKEK IDQLQEELLH TQLKYQRILE RLEKENKELR KLVLQKDDKG IHHRKLKKSL IDMYSEVLDV LSDYDASYN TQDHLPRVVV VGDQSAGKTS VLEMIAQARI FPRGSGEMMT RSPVKVTLSE GPHHVALFKD SSREFDLTKE E DLAALRHE ...String: ATDRGSESDK HFRKVSDKEK IDQLQEELLH TQLKYQRILE RLEKENKELR KLVLQKDDKG IHHRKLKKSL IDMYSEVLDV LSDYDASYN TQDHLPRVVV VGDQSAGKTS VLEMIAQARI FPRGSGEMMT RSPVKVTLSE GPHHVALFKD SSREFDLTKE E DLAALRHE IELRMRKNVK EGCTVSPETI SLNVKGPGLQ RMVLVDLPGV INTVTSGMAP DTKETIFSIS KAYMQNPNAI IL CIQDGSV DAERSIVTDL VSQMDPHGRR TIFVLTKVDL AEKNVASPSR IQQIIEGKLF PMKALGYFAV VTGKGNSSES IEA IREYEE EFFQNSKLLK TSMLKAHQVT TRNLSLAVSD CFWKMVRESV EQQADSFKAT RFNLETEWKN NYPRLRELDR NELF EKAKN EILDEVISLS QVTPKHWEEI LQQSLWERVS THVIENIYLP AAQTMNSGTF NTTVDIKLKQ WTDKQLPNKA VEVAW ETLQ EEFSRFMTEP KGKEHDDIFD KLKEAVKEES IKRHKWNDFA EDSLRVIQHN ALEDRSISDK QQWDAAIYFM EEALQA RLK DTENAIENMV GPDWKKRWLY WKNRTQEQCV HNETKNELEK MLKCNEEHPA YLASDEITTV RKNLESRGVE VDPSLIK DT WHQVYRRHFL KTALNHCNLC RRGFYYYQRH FVDSELECND VVLFWRIQRM LAITANTLRQ QLTNTEVRRL EKNVKEVL E DFAEDGEKKI KLLTGKRVQL AEDLKKVREI QEKLDAFIEA LHQEK UniProtKB: Dynamin-like GTPase OPA1, mitochondrial |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | helical array |
- Sample preparation
Sample preparation
| Buffer | pH: 7.2 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | TFS GLACIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 24.86 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.4 µm / Nominal defocus min: 0.6 µm |
- Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT |
|---|---|
| Output model |  PDB-8ef7:  PDB-8efs:  PDB-8eft: |
 Movie
Movie Controller
Controller




















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)