[English] 日本語
 Yorodumi
Yorodumi- EMDB-40192: CryoEM map of the locally refined soluble OPA1 Z-clip from the GD... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | CryoEM map of the locally refined soluble OPA1 Z-clip from the GDP-AlFx bound helical assembly on a lipid membrane | |||||||||
 Map data Map data | CryoEM map of the OPA1 Z-clip from the GDP-AlFx bound helical assembly on a lipid membrane | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | GTPase / Dynamin-family protein / mitochondrial fusion protein / mitochondria / Optic Atrophy / LIPID BINDING PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationRegulation of Apoptosis / mitochondrial inner membrane fusion / membrane tubulation / membrane bending activity / inner mitochondrial membrane organization / GTPase-dependent fusogenic activity / dynamin GTPase / cristae formation / peroxisome fission / : ...Regulation of Apoptosis / mitochondrial inner membrane fusion / membrane tubulation / membrane bending activity / inner mitochondrial membrane organization / GTPase-dependent fusogenic activity / dynamin GTPase / cristae formation / peroxisome fission / : / phosphatidic acid binding / cardiolipin binding / mitochondrial fission / GTP metabolic process / mitochondrial fusion / axonal transport of mitochondrion / negative regulation of release of cytochrome c from mitochondria / mitochondrial crista / intracellular distribution of mitochondria / positive regulation of interleukin-17 production / protein complex oligomerization / positive regulation of T-helper 17 cell lineage commitment / negative regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway / axon cytoplasm / visual perception / Mitochondrial protein degradation / mitochondrion organization / neural tube closure / mitochondrial intermembrane space / mitochondrial membrane / endocytosis / cellular senescence / microtubule binding / microtubule / mitochondrial outer membrane / mitochondrial inner membrane / GTPase activity / apoptotic process / dendrite / negative regulation of apoptotic process / GTP binding / magnesium ion binding / mitochondrion / nucleoplasm / membrane / cytosol / cytoplasm Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
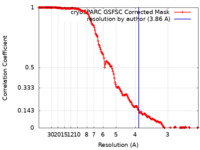
| Method | helical reconstruction / cryo EM / Resolution: 3.86 Å | |||||||||
 Authors Authors | Nyenhuis SB / Wu X / Strub MP / Yim YI / Stanton AE / Baena V / Syed ZA / Canagarajah B / Hammer JA / Hinshaw JE | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Nature / Year: 2023 Journal: Nature / Year: 2023Title: OPA1 helical structures give perspective to mitochondrial dysfunction. Authors: Sarah B Nyenhuis / Xufeng Wu / Marie-Paule Strub / Yang-In Yim / Abigail E Stanton / Valentina Baena / Zulfeqhar A Syed / Bertram Canagarajah / John A Hammer / Jenny E Hinshaw /  Abstract: Dominant optic atrophy is one of the leading causes of childhood blindness. Around 60-80% of cases are caused by mutations of the gene that encodes optic atrophy protein 1 (OPA1), a protein that has ...Dominant optic atrophy is one of the leading causes of childhood blindness. Around 60-80% of cases are caused by mutations of the gene that encodes optic atrophy protein 1 (OPA1), a protein that has a key role in inner mitochondrial membrane fusion and remodelling of cristae and is crucial for the dynamic organization and regulation of mitochondria. Mutations in OPA1 result in the dysregulation of the GTPase-mediated fusion process of the mitochondrial inner and outer membranes. Here we used cryo-electron microscopy methods to solve helical structures of OPA1 assembled on lipid membrane tubes, in the presence and absence of nucleotide. These helical assemblies organize into densely packed protein rungs with minimal inter-rung connectivity, and exhibit nucleotide-dependent dimerization of the GTPase domains-a hallmark of the dynamin superfamily of proteins. OPA1 also contains several unique secondary structures in the paddle domain that strengthen its membrane association, including membrane-inserting helices. The structural features identified in this study shed light on the effects of pathogenic point mutations on protein folding, inter-protein assembly and membrane interactions. Furthermore, mutations that disrupt the assembly interfaces and membrane binding of OPA1 cause mitochondrial fragmentation in cell-based assays, providing evidence of the biological relevance of these interactions. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_40192.map.gz emd_40192.map.gz | 234.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-40192-v30.xml emd-40192-v30.xml emd-40192.xml emd-40192.xml | 19.6 KB 19.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_40192_fsc.xml emd_40192_fsc.xml | 18.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_40192.png emd_40192.png | 84.7 KB | ||
| Masks |  emd_40192_msk_1.map emd_40192_msk_1.map | 476.8 MB |  Mask map Mask map | |
| Others |  emd_40192_additional_1.map.gz emd_40192_additional_1.map.gz emd_40192_additional_2.map.gz emd_40192_additional_2.map.gz emd_40192_half_map_1.map.gz emd_40192_half_map_1.map.gz emd_40192_half_map_2.map.gz emd_40192_half_map_2.map.gz | 423.4 MB 448.9 MB 442.8 MB 442.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-40192 http://ftp.pdbj.org/pub/emdb/structures/EMD-40192 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-40192 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-40192 | HTTPS FTP |
-Validation report
| Summary document |  emd_40192_validation.pdf.gz emd_40192_validation.pdf.gz | 1.2 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_40192_full_validation.pdf.gz emd_40192_full_validation.pdf.gz | 1.2 MB | Display | |
| Data in XML |  emd_40192_validation.xml.gz emd_40192_validation.xml.gz | 25.1 KB | Display | |
| Data in CIF |  emd_40192_validation.cif.gz emd_40192_validation.cif.gz | 32.9 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-40192 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-40192 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-40192 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-40192 | HTTPS FTP |
-Related structure data
| Related structure data |  8eewC  8ef7C  8effC  8efrC  8efsC  8eftC C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_40192.map.gz / Format: CCP4 / Size: 476.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_40192.map.gz / Format: CCP4 / Size: 476.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | CryoEM map of the OPA1 Z-clip from the GDP-AlFx bound helical assembly on a lipid membrane | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.2018 Å | ||||||||||||||||||||||||||||||||||||
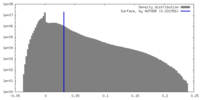
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_40192_msk_1.map emd_40192_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
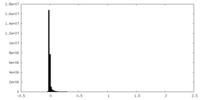
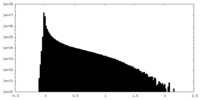
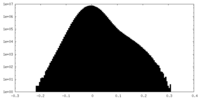
| Density Histograms |
-Additional map: CryoEM sharpened map of the OPA1 Z-clip from...
| File | emd_40192_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | CryoEM sharpened map of the OPA1 Z-clip from the GDP-AlFx bound helical assembly on a lipid membrane | ||||||||||||
| Projections & Slices |
| ||||||||||||
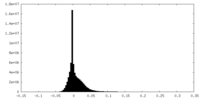
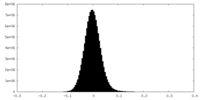
| Density Histograms |
-Additional map: CryoEM sharpened map 2 of the OPA1 Z-clip...
| File | emd_40192_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | CryoEM sharpened map 2 of the OPA1 Z-clip from the GDP-AlFx bound helical assembly on a lipid membrane | ||||||||||||
| Projections & Slices |
| ||||||||||||
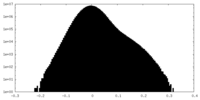
| Density Histograms |
-Half map: CryoEM half-map B of the OPA1 Z-clip from...
| File | emd_40192_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | CryoEM half-map B of the OPA1 Z-clip from the GDP-AlFx bound helical assembly on a lipid membrane | ||||||||||||
| Projections & Slices |
| ||||||||||||
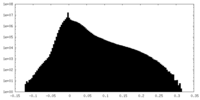
| Density Histograms |
-Half map: CryoEM half-map A of the OPA1 Z-clip from...
| File | emd_40192_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | CryoEM half-map A of the OPA1 Z-clip from the GDP-AlFx bound helical assembly on a lipid membrane | ||||||||||||
| Projections & Slices |
| ||||||||||||
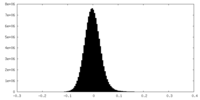
| Density Histograms |
- Sample components
Sample components
-Entire : sOPA1 helical assembly on a lipid membrane
| Entire | Name: sOPA1 helical assembly on a lipid membrane |
|---|---|
| Components |
|
-Supramolecule #1: sOPA1 helical assembly on a lipid membrane
| Supramolecule | Name: sOPA1 helical assembly on a lipid membrane / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: sOPA1 protein
| Macromolecule | Name: sOPA1 protein / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  |
| Sequence | String: ATDRGSESDK HFRKVSDKEK IDQLQEELLH TQLKYQRILE RLEKENKELR KLVLQKDDKG IHHRKLKKSL IDMYSEVLD VLSDYDASYN TQDHLPRVVV VGDQSAGKTS VLEMIAQARI FPRGSGEMMT RSPVKVTLSE G PHHVALFK DSSREFDLTK EEDLAALRHE ...String: ATDRGSESDK HFRKVSDKEK IDQLQEELLH TQLKYQRILE RLEKENKELR KLVLQKDDKG IHHRKLKKSL IDMYSEVLD VLSDYDASYN TQDHLPRVVV VGDQSAGKTS VLEMIAQARI FPRGSGEMMT RSPVKVTLSE G PHHVALFK DSSREFDLTK EEDLAALRHE IELRMRKNVK EGCTVSPETI SLNVKGPGLQ RMVLVDLPGV IN TVTSGMA PDTKETIFSI SKAYMQNPNA IILCIQDGSV DAERSIVTDL VSQMDPHGRR TIFVLTKVDL AEK NVASPS RIQQIIEGKL FPMKALGYFA VVTGKGNSSE SIEAIREYEE EFFQNSKLLK TSMLKAHQVT TRNL SLAVS DCFWKMVRES VEQQADSFKA TRFNLETEWK NNYPRLRELD RNELFEKAKN EILDEVISLS QVTPK HWEE ILQQSLWERV STHVIENIYL PAAQTMNSGT FNTTVDIKLK QWTDKQLPNK AVEVAWETLQ EEFSRF MTE PKGKEHDDIF DKLKEAVKEE SIKRHKWNDF AEDSLRVIQH NALEDRSISD KQQWDAAIYF MEEALQA RL KDTENAIENM VGPDWKKRWL YWKNRTQEQC VHNETKNELE KMLKCNEEHP AYLASDEITT VRKNLESR G VEVDPSLIKD TWHQVYRRHF LKTALNHCNL CRRGFYYYQR HFVDSELECN DVVLFWRIQR MLAITANTL RQQLTNTEVR RLEKNVKEVL EDFAEDGEKK IKLLTGKRVQ LAEDLKKVRE IQEKLDAFIE ALHQEK UniProtKB: Dynamin-like GTPase OPA1, mitochondrial |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | helical array |
- Sample preparation
Sample preparation
| Buffer | pH: 7.2 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.4 µm / Nominal defocus min: 0.3 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT |
|---|
 Movie
Movie Controller
Controller




















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)