+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Pseudomonas phage phiKZ PhuZ Filament | ||||||||||||
 Map data Map data | unsharpened and filtered to the FSC0.143 cutoff | ||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | bacteriophage / structural protein / viral tubulin / VIRAL PROTEIN | ||||||||||||
| Biological species |  Pseudomonas phage phiKZ (virus) Pseudomonas phage phiKZ (virus) | ||||||||||||
| Method | subtomogram averaging / cryo EM / Resolution: 22.0 Å | ||||||||||||
 Authors Authors | Laughlin TG / Villa E | ||||||||||||
| Funding support |  United States, 3 items United States, 3 items
| ||||||||||||
 Citation Citation |  Journal: bioRxiv / Year: 2023 Journal: bioRxiv / Year: 2023Title: Identifying the core genome of the nucleus-forming bacteriophage family and characterization of phage RAY. Abstract: We recently discovered that some bacteriophages establish a nucleus-like replication compartment (phage nucleus), but the core genes that define nucleus-based phage replication and their phylogenetic ...We recently discovered that some bacteriophages establish a nucleus-like replication compartment (phage nucleus), but the core genes that define nucleus-based phage replication and their phylogenetic distribution were unknown. By studying phages that encode the major phage nucleus protein chimallin, including previously sequenced yet uncharacterized phages, we discovered that chimallin-encoding phages share a set of 72 highly conserved genes encoded within seven distinct gene blocks. Of these, 21 core genes are unique to this group, and all but one of these unique genes encode proteins of unknown function. We propose that phages with this core genome comprise a novel viral family we term Chimalliviridae. Fluorescence microscopy and cryo-electron tomography studies of phage vB_EamM_RAY confirm that many of the key steps of nucleus-based replication encoded in the core genome are conserved among diverse chimalliviruses, and reveal that non-core components can confer intriguing variations on this replication mechanism. For instance, unlike previously studied nucleus-forming phages, RAY doesn't degrade the host genome, and its PhuZ homolog appears to form a five-stranded filament with a lumen. This work expands our understanding of phage nucleus and PhuZ spindle diversity and function, providing a roadmap for identifying key mechanisms underlying nucleus-based phage replication. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_28010.map.gz emd_28010.map.gz | 298.7 KB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-28010-v30.xml emd-28010-v30.xml emd-28010.xml emd-28010.xml | 17.7 KB 17.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_28010_fsc.xml emd_28010_fsc.xml | 1.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_28010.png emd_28010.png | 25.1 KB | ||
| Masks |  emd_28010_msk_1.map emd_28010_msk_1.map | 333.8 KB |  Mask map Mask map | |
| Filedesc metadata |  emd-28010.cif.gz emd-28010.cif.gz | 4.7 KB | ||
| Others |  emd_28010_half_map_1.map.gz emd_28010_half_map_1.map.gz emd_28010_half_map_2.map.gz emd_28010_half_map_2.map.gz | 248.8 KB 248.2 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-28010 http://ftp.pdbj.org/pub/emdb/structures/EMD-28010 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-28010 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-28010 | HTTPS FTP |
-Related structure data
| Related structure data | C: citing same article ( |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_28010.map.gz / Format: CCP4 / Size: 333 KB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_28010.map.gz / Format: CCP4 / Size: 333 KB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | unsharpened and filtered to the FSC0.143 cutoff | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 3.457 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_28010_msk_1.map emd_28010_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_28010_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_28010_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Pseudomonas phage phiKZ PhuZ Filament
| Entire | Name: Pseudomonas phage phiKZ PhuZ Filament |
|---|---|
| Components |
|
-Supramolecule #1: Pseudomonas phage phiKZ PhuZ Filament
| Supramolecule | Name: Pseudomonas phage phiKZ PhuZ Filament / type: complex / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:  Pseudomonas phage phiKZ (virus) Pseudomonas phage phiKZ (virus) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | subtomogram averaging |
| Aggregation state | cell |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Grid | Model: Quantifoil R2/1 / Material: COPPER / Mesh: 200 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 60 sec. / Pretreatment - Atmosphere: OTHER / Pretreatment - Pressure: 0.019 kPa / Details: 20 mA in a PELCO easiGLO |
| Vitrification | Cryogen name: ETHANE-PROPANE / Instrument: HOMEMADE PLUNGER |
| Details | cell suspension |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Quantum LS / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Number grids imaged: 2 / Average electron dose: 2.5 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 6.0 µm / Nominal defocus min: 4.0 µm / Nominal magnification: 42000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller












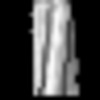
 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)













































