+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | CryoEM structure of miniGo-coupled hM4Di in complex with DCZ | |||||||||
 Map data Map data | CryoEM structure of miniGo-coupled hM4Di in complex with DCZ | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | GPCR / DCZ / active state / MEMBRANE PROTEIN / hM4Di / DREADD | |||||||||
| Function / homology |  Function and homology information Function and homology informationMuscarinic acetylcholine receptors / G protein-coupled acetylcholine receptor activity / adenylate cyclase-inhibiting G protein-coupled acetylcholine receptor signaling pathway / mu-type opioid receptor binding / corticotropin-releasing hormone receptor 1 binding / vesicle docking involved in exocytosis / G protein-coupled dopamine receptor signaling pathway / regulation of locomotion / regulation of heart contraction / parallel fiber to Purkinje cell synapse ...Muscarinic acetylcholine receptors / G protein-coupled acetylcholine receptor activity / adenylate cyclase-inhibiting G protein-coupled acetylcholine receptor signaling pathway / mu-type opioid receptor binding / corticotropin-releasing hormone receptor 1 binding / vesicle docking involved in exocytosis / G protein-coupled dopamine receptor signaling pathway / regulation of locomotion / regulation of heart contraction / parallel fiber to Purkinje cell synapse / G protein-coupled receptor signaling pathway, coupled to cyclic nucleotide second messenger / postsynaptic modulation of chemical synaptic transmission / G protein-coupled serotonin receptor binding / adenylate cyclase regulator activity / adenylate cyclase-inhibiting serotonin receptor signaling pathway / muscle contraction / locomotory behavior / negative regulation of insulin secretion / GABA-ergic synapse / adenylate cyclase-modulating G protein-coupled receptor signaling pathway / G-protein beta/gamma-subunit complex binding / Olfactory Signaling Pathway / Activation of the phototransduction cascade / G beta:gamma signalling through PLC beta / Presynaptic function of Kainate receptors / Thromboxane signalling through TP receptor / G protein-coupled acetylcholine receptor signaling pathway / Activation of G protein gated Potassium channels / Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits / G-protein activation / G beta:gamma signalling through CDC42 / Prostacyclin signalling through prostacyclin receptor / Glucagon signaling in metabolic regulation / G beta:gamma signalling through BTK / Synthesis, secretion, and inactivation of Glucagon-like Peptide-1 (GLP-1) / ADP signalling through P2Y purinoceptor 12 / photoreceptor disc membrane / Glucagon-type ligand receptors / Sensory perception of sweet, bitter, and umami (glutamate) taste / Adrenaline,noradrenaline inhibits insulin secretion / Vasopressin regulates renal water homeostasis via Aquaporins / Glucagon-like Peptide-1 (GLP1) regulates insulin secretion / G alpha (z) signalling events / ADP signalling through P2Y purinoceptor 1 / ADORA2B mediated anti-inflammatory cytokines production / cellular response to catecholamine stimulus / G beta:gamma signalling through PI3Kgamma / adenylate cyclase-activating dopamine receptor signaling pathway / Cooperation of PDCL (PhLP1) and TRiC/CCT in G-protein beta folding / GPER1 signaling / G-protein beta-subunit binding / cellular response to prostaglandin E stimulus / heterotrimeric G-protein complex / G alpha (12/13) signalling events / Inactivation, recovery and regulation of the phototransduction cascade / extracellular vesicle / sensory perception of taste / Thrombin signalling through proteinase activated receptors (PARs) / signaling receptor complex adaptor activity / retina development in camera-type eye / cell body / G protein activity / GTPase binding / presynaptic membrane / Ca2+ pathway / fibroblast proliferation / High laminar flow shear stress activates signaling by PIEZO1 and PECAM1:CDH5:KDR in endothelial cells / G alpha (i) signalling events / G alpha (s) signalling events / phospholipase C-activating G protein-coupled receptor signaling pathway / G alpha (q) signalling events / Hydrolases; Acting on acid anhydrides; Acting on GTP to facilitate cellular and subcellular movement / chemical synaptic transmission / Ras protein signal transduction / postsynaptic membrane / cell surface receptor signaling pathway / Extra-nuclear estrogen signaling / cell population proliferation / G protein-coupled receptor signaling pathway / lysosomal membrane / GTPase activity / synapse / dendrite / GTP binding / protein-containing complex binding / glutamatergic synapse / signal transduction / extracellular exosome / metal ion binding / membrane / plasma membrane / cytoplasm / cytosol Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /  | |||||||||
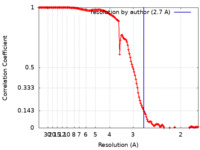
| Method | single particle reconstruction / cryo EM / Resolution: 2.7 Å | |||||||||
 Authors Authors | Zhang S / Fay JF / Roth BL | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Nature / Year: 2022 Journal: Nature / Year: 2022Title: Molecular basis for selective activation of DREADD-based chemogenetics. Authors: Shicheng Zhang / Ryan H Gumpper / Xi-Ping Huang / Yongfeng Liu / Brian E Krumm / Can Cao / Jonathan F Fay / Bryan L Roth /  Abstract: Designer receptors exclusively activated by designer drugs (DREADDs) represent a powerful chemogenetic technology for the remote control of neuronal activity and cellular signalling. The muscarinic ...Designer receptors exclusively activated by designer drugs (DREADDs) represent a powerful chemogenetic technology for the remote control of neuronal activity and cellular signalling. The muscarinic receptor-based DREADDs are the most widely used chemogenetic tools in neuroscience research. The G-coupled DREADD (hM3Dq) is used to enhance neuronal activity, whereas the G-coupled DREADD (hM4Di) is utilized to inhibit neuronal activity. Here we report four DREADD-related cryogenic electron microscopy high-resolution structures: a hM3Dq-miniG complex and a hM4Di-miniG complex bound to deschloroclozapine; a hM3Dq-miniG complex bound to clozapine-N-oxide; and a hM3R-miniG complex bound to iperoxo. Complemented with mutagenesis, functional and computational simulation data, our structures reveal key details of the recognition of DREADD chemogenetic actuators and the molecular basis for activation. These findings should accelerate the structure-guided discovery of next-generation chemogenetic tools. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_27967.map.gz emd_27967.map.gz | 80.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-27967-v30.xml emd-27967-v30.xml emd-27967.xml emd-27967.xml | 27.3 KB 27.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_27967_fsc.xml emd_27967_fsc.xml | 13.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_27967.png emd_27967.png | 62.3 KB | ||
| Filedesc metadata |  emd-27967.cif.gz emd-27967.cif.gz | 7.3 KB | ||
| Others |  emd_27967_additional_1.map.gz emd_27967_additional_1.map.gz emd_27967_half_map_1.map.gz emd_27967_half_map_1.map.gz emd_27967_half_map_2.map.gz emd_27967_half_map_2.map.gz | 84.9 MB 84.6 MB 84.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-27967 http://ftp.pdbj.org/pub/emdb/structures/EMD-27967 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27967 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27967 | HTTPS FTP |
-Related structure data
| Related structure data |  8e9xMC  8e9wC  8e9yC  8e9zC  8ea0C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_27967.map.gz / Format: CCP4 / Size: 91.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_27967.map.gz / Format: CCP4 / Size: 91.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | CryoEM structure of miniGo-coupled hM4Di in complex with DCZ | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.88 Å | ||||||||||||||||||||||||||||||||||||
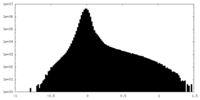
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: Additional Map
| File | emd_27967_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Additional Map | ||||||||||||
| Projections & Slices |
| ||||||||||||
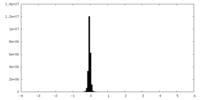
| Density Histograms |
-Half map: Half Map 1
| File | emd_27967_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half Map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
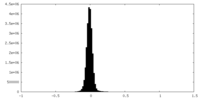
| Density Histograms |
-Half map: Half Map 2
| File | emd_27967_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half Map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
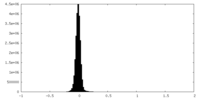
| Density Histograms |
- Sample components
Sample components
-Entire : Gq-coupled hM3R complex
| Entire | Name: Gq-coupled hM3R complex |
|---|---|
| Components |
|
-Supramolecule #1: Gq-coupled hM3R complex
| Supramolecule | Name: Gq-coupled hM3R complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#5 |
|---|
-Supramolecule #2: muscarinic acetylcholine receptor 4, miniGo protein, Guanine nucl...
| Supramolecule | Name: muscarinic acetylcholine receptor 4, miniGo protein, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1-#4 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Supramolecule #3: Single-chain variable fragment scFv16
| Supramolecule | Name: Single-chain variable fragment scFv16 / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #5 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Muscarinic acetylcholine receptor M4
| Macromolecule | Name: Muscarinic acetylcholine receptor M4 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 39.225297 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MANFTPVNGS SGNQSVRLVT SSSHNRYETV EMVFIATVTG SLSLVTVVGN ILVMLSIKVN RQLQTVNNYF LFSLACADLI IGAFSMNLY TVYIIKGYWP LGAVVCDLWL ALDCVVSNAS VMNLLIISFD RYFCVTKPLT YPARRTTKMA GLMIAAAWVL S FVLWAPAI ...String: MANFTPVNGS SGNQSVRLVT SSSHNRYETV EMVFIATVTG SLSLVTVVGN ILVMLSIKVN RQLQTVNNYF LFSLACADLI IGAFSMNLY TVYIIKGYWP LGAVVCDLWL ALDCVVSNAS VMNLLIISFD RYFCVTKPLT YPARRTTKMA GLMIAAAWVL S FVLWAPAI LFWQFVVGKR TVPDNQCFIQ FLSNPAVTFG TAIAGFYLPV VIMTVLYIHI SLASRSRVHK HRPEGPKEKK AK TLARKFA SIARNQVRKK RQMAARERKV TRTIFAILLA FILTWTPYNV MVLVNTFCQS CIPDTVWSIG YWLCYVNSTI NPA CYALCN ATFKKTFRHL LLCQYRNIGT AR UniProtKB: Muscarinic acetylcholine receptor M4, Muscarinic acetylcholine receptor M4 |
-Macromolecule #2: miniGo
| Macromolecule | Name: miniGo / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 25.159777 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: TLSAEDKAAV ERSKMIEKNL KEDGISAAKD VKLLLLGADN SGKSTIVKQM KIIHGGSGGS GGTTGIVETH FTFKNLHFRL FDVGGQRSE RKKWIHCFED VTAIIFCVDL SDYNRMHESL MLFDSICNNK FFIDTSIILF LNKKDLFGEK IKKSPLTICF P EYTGPNTY ...String: TLSAEDKAAV ERSKMIEKNL KEDGISAAKD VKLLLLGADN SGKSTIVKQM KIIHGGSGGS GGTTGIVETH FTFKNLHFRL FDVGGQRSE RKKWIHCFED VTAIIFCVDL SDYNRMHESL MLFDSICNNK FFIDTSIILF LNKKDLFGEK IKKSPLTICF P EYTGPNTY EDAAAYIQAQ FESKNRSPNK EIYCHMTCAT DTNNAQVIFD AVTDIIIANN LRGCGLY UniProtKB: Guanine nucleotide-binding protein G(o) subunit alpha, Guanine nucleotide-binding protein G(o) subunit alpha |
-Macromolecule #3: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 37.285734 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: SELDQLRQEA EQLKNQIRDA RKACADATLS QITNNIDPVG RIQMRTRRTL RGHLAKIYAM HWGTDSRLLV SASQDGKLII WDSYTTNKV HAIPLRSSWV MTCAYAPSGN YVACGGLDNI CSIYNLKTRE GNVRVSRELA GHTGYLSCCR FLDDNQIVTS S GDTTCALW ...String: SELDQLRQEA EQLKNQIRDA RKACADATLS QITNNIDPVG RIQMRTRRTL RGHLAKIYAM HWGTDSRLLV SASQDGKLII WDSYTTNKV HAIPLRSSWV MTCAYAPSGN YVACGGLDNI CSIYNLKTRE GNVRVSRELA GHTGYLSCCR FLDDNQIVTS S GDTTCALW DIETGQQTTT FTGHTGDVMS LSLAPDTRLF VSGACDASAK LWDVREGMCR QTFTGHESDI NAICFFPNGN AF ATGSDDA TCRLFDLRAD QELMTYSHDN IICGITSVSF SKSGRLLLAG YDDFNCNVWD ALKADRAGVL AGHDNRVSCL GVT DDGMAV ATGSWDSFLK IWN UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 |
-Macromolecule #4: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 7.861143 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MASNNTASIA QARKLVEQLK MEANIDRIKV SKAAADLMAY CEAHAKEDPL LTPVPASENP FREKKFFCAI L UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 |
-Macromolecule #5: scFv16
| Macromolecule | Name: scFv16 / type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 26.679721 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: DVQLVESGGG LVQPGGSRKL SCSASGFAFS SFGMHWVRQA PEKGLEWVAY ISSGSGTIYY ADTVKGRFTI SRDDPKNTLF LQMTSLRSE DTAMYYCVRS IYYYGSSPFD FWGQGTTLTV SSGGGGSGGG GSGGGGSDIV MTQATSSVPV TPGESVSISC R SSKSLLHS ...String: DVQLVESGGG LVQPGGSRKL SCSASGFAFS SFGMHWVRQA PEKGLEWVAY ISSGSGTIYY ADTVKGRFTI SRDDPKNTLF LQMTSLRSE DTAMYYCVRS IYYYGSSPFD FWGQGTTLTV SSGGGGSGGG GSGGGGSDIV MTQATSSVPV TPGESVSISC R SSKSLLHS NGNTYLYWFL QRPGQSPQLL IYRMSNLASG VPDRFSGSGS GTAFTLTISR LEAEDVGVYY CMQHLEYPLT FG AGTKLEL KAAA |
-Macromolecule #6: 11-(4-methylpiperazin-1-yl)-5H-dibenzo[b,e][1,4]diazepine
| Macromolecule | Name: 11-(4-methylpiperazin-1-yl)-5H-dibenzo[b,e][1,4]diazepine type: ligand / ID: 6 / Number of copies: 1 / Formula: WEC |
|---|---|
| Molecular weight | Theoretical: 292.378 Da |
| Chemical component information |  ChemComp-WEC: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE-PROPANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Number real images: 2982 / Average electron dose: 46.1 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.9 µm / Nominal defocus min: 0.2 µm |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller






























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)