+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Purification of Enterovirus A71, strain 4643, WT capsid | |||||||||
 Map data Map data | Purification of Enterovirus A71, strain 4643, WT capsid | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | enterovirus / thermostability / capsid / VIRUS | |||||||||
| Function / homology |  Function and homology information Function and homology informationsymbiont-mediated suppression of host cytoplasmic pattern recognition receptor signaling pathway via inhibition of MDA-5 activity / picornain 2A / symbiont-mediated suppression of host mRNA export from nucleus / symbiont genome entry into host cell via pore formation in plasma membrane / picornain 3C / T=pseudo3 icosahedral viral capsid / host cell cytoplasmic vesicle membrane / nucleoside-triphosphate phosphatase / channel activity / monoatomic ion transmembrane transport ...symbiont-mediated suppression of host cytoplasmic pattern recognition receptor signaling pathway via inhibition of MDA-5 activity / picornain 2A / symbiont-mediated suppression of host mRNA export from nucleus / symbiont genome entry into host cell via pore formation in plasma membrane / picornain 3C / T=pseudo3 icosahedral viral capsid / host cell cytoplasmic vesicle membrane / nucleoside-triphosphate phosphatase / channel activity / monoatomic ion transmembrane transport / DNA replication / RNA helicase activity / endocytosis involved in viral entry into host cell / symbiont-mediated activation of host autophagy / RNA-directed RNA polymerase / cysteine-type endopeptidase activity / viral RNA genome replication / RNA-directed RNA polymerase activity / DNA-templated transcription / virion attachment to host cell / host cell nucleus / structural molecule activity / ATP hydrolysis activity / proteolysis / RNA binding / zinc ion binding / ATP binding / membrane Similarity search - Function | |||||||||
| Biological species |   Enterovirus A71 / Enterovirus A71 /   Human enterovirus 71 Human enterovirus 71 | |||||||||
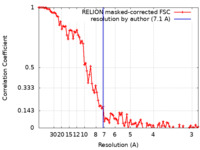
| Method | single particle reconstruction / cryo EM / Resolution: 7.1 Å | |||||||||
 Authors Authors | Catching A / Capponi S / Andino R | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2023 Journal: Nat Commun / Year: 2023Title: A tradeoff between enterovirus A71 particle stability and cell entry. Authors: Adam Catching / Ming Te Yeh / Simone Bianco / Sara Capponi / Raul Andino /  Abstract: A central role of viral capsids is to protect the viral genome from the harsh extracellular environment while facilitating initiation of infection when the virus encounters a target cell. Viruses are ...A central role of viral capsids is to protect the viral genome from the harsh extracellular environment while facilitating initiation of infection when the virus encounters a target cell. Viruses are thought to have evolved an optimal equilibrium between particle stability and efficiency of cell entry. In this study, we genetically perturb this equilibrium in a non-enveloped virus, enterovirus A71 to determine its structural basis. We isolate a single-point mutation variant with increased particle thermotolerance and decreased efficiency of cell entry. Using cryo-electron microscopy and molecular dynamics simulations, we determine that the thermostable native particles have acquired an expanded conformation that results in a significant increase in protein dynamics. Examining the intermediate states of the thermostable variant reveals a potential pathway for uncoating. We propose a sequential release of the lipid pocket factor, followed by internal VP4 and ultimately the viral RNA. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_27863.map.gz emd_27863.map.gz | 26.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-27863-v30.xml emd-27863-v30.xml emd-27863.xml emd-27863.xml | 19.7 KB 19.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_27863_fsc.xml emd_27863_fsc.xml | 9.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_27863.png emd_27863.png | 129.3 KB | ||
| Masks |  emd_27863_msk_1.map emd_27863_msk_1.map | 1.1 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-27863.cif.gz emd-27863.cif.gz | 6.3 KB | ||
| Others |  emd_27863_half_map_1.map.gz emd_27863_half_map_1.map.gz emd_27863_half_map_2.map.gz emd_27863_half_map_2.map.gz | 49.5 MB 49.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-27863 http://ftp.pdbj.org/pub/emdb/structures/EMD-27863 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27863 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27863 | HTTPS FTP |
-Validation report
| Summary document |  emd_27863_validation.pdf.gz emd_27863_validation.pdf.gz | 1.2 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_27863_full_validation.pdf.gz emd_27863_full_validation.pdf.gz | 1.2 MB | Display | |
| Data in XML |  emd_27863_validation.xml.gz emd_27863_validation.xml.gz | 16.3 KB | Display | |
| Data in CIF |  emd_27863_validation.cif.gz emd_27863_validation.cif.gz | 21.4 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-27863 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-27863 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-27863 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-27863 | HTTPS FTP |
-Related structure data
| Related structure data |  8e3cMC  8e2xC  8e2yC  8e31C  8e38C  8e39C  8e3aC  8e3bC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_27863.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_27863.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Purification of Enterovirus A71, strain 4643, WT capsid | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.44 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_27863_msk_1.map emd_27863_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Purification of Enterovirus A71, strain 4643, WT capsid
| File | emd_27863_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Purification of Enterovirus A71, strain 4643, WT capsid | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Purification of Enterovirus A71, strain 4643, WT capsid
| File | emd_27863_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Purification of Enterovirus A71, strain 4643, WT capsid | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Human enterovirus 71
| Entire | Name:   Human enterovirus 71 Human enterovirus 71 |
|---|---|
| Components |
|
-Supramolecule #1: Human enterovirus 71
| Supramolecule | Name: Human enterovirus 71 / type: virus / ID: 1 / Parent: 0 / Macromolecule list: all / NCBI-ID: 39054 / Sci species name: Human enterovirus 71 / Virus type: VIRUS-LIKE PARTICLE / Virus isolate: STRAIN / Virus enveloped: No / Virus empty: No |
|---|---|
| Host (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Virus shell | Shell ID: 1 / Name: Capsid / Diameter: 300.0 Å / T number (triangulation number): 1 |
-Macromolecule #1: VP1
| Macromolecule | Name: VP1 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO / EC number: picornain 2A |
|---|---|
| Source (natural) | Organism:   Enterovirus A71 / Strain: Tainan/4643/98 Enterovirus A71 / Strain: Tainan/4643/98 |
| Molecular weight | Theoretical: 25.366697 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: SHSTAETTLD SFFSRAGLVG EIDLPLEGTT NPNGYANWDI DITGYAQMRR KVELFTYMRF DAEFTFVACT PTGQVVPQLL QYMFVPPGA PEPDSRESLA WQTATNPSVF VKLSDPPAQV SVPFMSPASA YQWFYDGYPT FGEHKQEKDL EYGACPNNMM G TFSVRTVG ...String: SHSTAETTLD SFFSRAGLVG EIDLPLEGTT NPNGYANWDI DITGYAQMRR KVELFTYMRF DAEFTFVACT PTGQVVPQLL QYMFVPPGA PEPDSRESLA WQTATNPSVF VKLSDPPAQV SVPFMSPASA YQWFYDGYPT FGEHKQEKDL EYGACPNNMM G TFSVRTVG TSKSKYPLVI RIYMRMKHVR AWIPRPMRNQ NYLFKANPNY AGNFIKPTGA SRTAITT UniProtKB: Genome polyprotein |
-Macromolecule #2: VP2
| Macromolecule | Name: VP2 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Enterovirus A71 / Strain: Tainan/4643/98 Enterovirus A71 / Strain: Tainan/4643/98 |
| Molecular weight | Theoretical: 25.803088 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: LTIGNSTITT QEAANIIVGY GEWPSYCSDS DATAVDKPTR PDVSVNRFYT LDTKLWEKSS KGWYWKFPDV LTETGVFGQN AQFHYLYRS GFCIHVQCNA SKFHQGALLV AVLPEYVIGT VAGGTGTEDS HPPYKQTQPG ADGFELQHPY VLDAGIPISQ L TVCPHQWI ...String: LTIGNSTITT QEAANIIVGY GEWPSYCSDS DATAVDKPTR PDVSVNRFYT LDTKLWEKSS KGWYWKFPDV LTETGVFGQN AQFHYLYRS GFCIHVQCNA SKFHQGALLV AVLPEYVIGT VAGGTGTEDS HPPYKQTQPG ADGFELQHPY VLDAGIPISQ L TVCPHQWI NLRTNNCATI IVPYINALPF DSALNHCNFG LLVVPISPLD YDQGATPVIP ITITLAPMCS EFAGLRQ UniProtKB: Genome polyprotein |
-Macromolecule #3: VP3
| Macromolecule | Name: VP3 / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Enterovirus A71 / Strain: Tainan/4643/98 Enterovirus A71 / Strain: Tainan/4643/98 |
| Molecular weight | Theoretical: 25.811473 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: GFPTELKPGT NQFLTTDDGV SAPILPNFHP TPCIHIPGEV RNLLELCQVE TILEVNNVPT NATSLMERLR FPVSAQAGKG ELCAVFRAD PGRSGPWQST LLGQLCGYYT QWSGSLEVTF MFTGSFMATG KMLIAYTPPG GPLPKDRATA MLGTHVIWDF G LQSSVTLV ...String: GFPTELKPGT NQFLTTDDGV SAPILPNFHP TPCIHIPGEV RNLLELCQVE TILEVNNVPT NATSLMERLR FPVSAQAGKG ELCAVFRAD PGRSGPWQST LLGQLCGYYT QWSGSLEVTF MFTGSFMATG KMLIAYTPPG GPLPKDRATA MLGTHVIWDF G LQSSVTLV IPWISNTHYR AHARDGVFDY YTTGLVSIWY QTNYVVPIGA PNTAYIIALA AAQKNFTMQL CKDASDIL UniProtKB: Genome polyprotein |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7 |
|---|---|
| Grid | Model: Quantifoil R2/1 / Material: COPPER / Mesh: 200 / Support film - Material: CARBON / Support film - topology: CONTINUOUS / Support film - Film thickness: 0.2 |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 297 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI ARCTICA |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Digitization - Dimensions - Width: 6000 pixel / Digitization - Dimensions - Height: 4000 pixel / Number grids imaged: 1 / Average exposure time: 6.0 sec. / Average electron dose: 64.1 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.5 µm / Nominal magnification: 45000 |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)