+ データを開く
データを開く
- 基本情報
基本情報
| 登録情報 |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| タイトル | Open state of RFC:PCNA bound to a 3' ss/dsDNA junction | ||||||||||||
 マップデータ マップデータ | Open RFC-PCNA | ||||||||||||
 試料 試料 |
| ||||||||||||
 キーワード キーワード | REPLICATION-DNA complex | ||||||||||||
| 機能・相同性 |  機能・相同性情報 機能・相同性情報positive regulation of DNA metabolic process / Rad17 RFC-like complex / Mismatch repair (MMR) directed by MSH2:MSH6 (MutSalpha) / Gap-filling DNA repair synthesis and ligation in GG-NER / meiotic mismatch repair / Processive synthesis on the lagging strand / Elg1 RFC-like complex / Removal of the Flap Intermediate / DNA replication factor C complex / Ctf18 RFC-like complex ...positive regulation of DNA metabolic process / Rad17 RFC-like complex / Mismatch repair (MMR) directed by MSH2:MSH6 (MutSalpha) / Gap-filling DNA repair synthesis and ligation in GG-NER / meiotic mismatch repair / Processive synthesis on the lagging strand / Elg1 RFC-like complex / Removal of the Flap Intermediate / DNA replication factor C complex / Ctf18 RFC-like complex / Polymerase switching / E3 ubiquitin ligases ubiquitinate target proteins / maintenance of DNA trinucleotide repeats / DNA clamp loader activity / SUMOylation of DNA replication proteins / Translesion synthesis by REV1 / Translesion synthesis by POLK / Translesion synthesis by POLI / Translesion Synthesis by POLH / DNA replication checkpoint signaling / establishment of mitotic sister chromatid cohesion / Termination of translesion DNA synthesis / Activation of ATR in response to replication stress / PCNA complex / lagging strand elongation / DNA damage tolerance / silent mating-type cassette heterochromatin formation / sister chromatid cohesion / mitotic sister chromatid cohesion / error-free translesion synthesis / DNA polymerase processivity factor activity / leading strand elongation / Gap-filling DNA repair synthesis and ligation in TC-NER / Dual incision in TC-NER / subtelomeric heterochromatin formation / translesion synthesis / mismatch repair / positive regulation of DNA repair / DNA damage checkpoint signaling / replication fork / positive regulation of DNA replication / nucleotide-excision repair / DNA-templated DNA replication / mitotic cell cycle / chromosome, telomeric region / cell division / DNA repair / ATP hydrolysis activity / DNA binding / ATP binding / identical protein binding / nucleus / cytosol 類似検索 - 分子機能 | ||||||||||||
| 生物種 |  | ||||||||||||
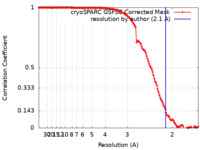
| 手法 | 単粒子再構成法 / クライオ電子顕微鏡法 / 解像度: 2.1 Å | ||||||||||||
 データ登録者 データ登録者 | Schrecker M / Hite RK | ||||||||||||
| 資金援助 |  米国, 3件 米国, 3件
| ||||||||||||
 引用 引用 |  ジャーナル: Elife / 年: 2022 ジャーナル: Elife / 年: 2022タイトル: Multistep loading of a DNA sliding clamp onto DNA by replication factor C. 著者: Marina Schrecker / Juan C Castaneda / Sujan Devbhandari / Charanya Kumar / Dirk Remus / Richard K Hite /  要旨: The DNA sliding clamp proliferating cell nuclear antigen (PCNA) is an essential co-factor for many eukaryotic DNA metabolic enzymes. PCNA is loaded around DNA by the ATP-dependent clamp loader ...The DNA sliding clamp proliferating cell nuclear antigen (PCNA) is an essential co-factor for many eukaryotic DNA metabolic enzymes. PCNA is loaded around DNA by the ATP-dependent clamp loader replication factor C (RFC), which acts at single-stranded (ss)/double-stranded DNA (dsDNA) junctions harboring a recessed 3' end (3' ss/dsDNA junctions) and at DNA nicks. To illuminate the loading mechanism we have investigated the structure of RFC:PCNA bound to ATPγS and 3' ss/dsDNA junctions or nicked DNA using cryogenic electron microscopy. Unexpectedly, we observe open and closed PCNA conformations in the RFC:PCNA:DNA complex, revealing that PCNA can adopt an open, planar conformation that allows direct insertion of dsDNA, and raising the question of whether PCNA ring closure is mechanistically coupled to ATP hydrolysis. By resolving multiple DNA-bound states of RFC:PCNA we observe that partial melting facilitates lateral insertion into the central channel formed by RFC:PCNA. We also resolve the Rfc1 N-terminal domain and demonstrate that its single BRCT domain participates in coordinating DNA prior to insertion into the central RFC channel, which promotes PCNA loading on the lagging strand of replication forks in vitro. Combined, our data suggest a comprehensive and fundamentally revised model for the RFC-catalyzed loading of PCNA onto DNA. | ||||||||||||
| 履歴 |
|
- 構造の表示
構造の表示
| 添付画像 |
|---|
- ダウンロードとリンク
ダウンロードとリンク
-EMDBアーカイブ
| マップデータ |  emd_27663.map.gz emd_27663.map.gz | 109 MB |  EMDBマップデータ形式 EMDBマップデータ形式 | |
|---|---|---|---|---|
| ヘッダ (付随情報) |  emd-27663-v30.xml emd-27663-v30.xml emd-27663.xml emd-27663.xml | 31 KB 31 KB | 表示 表示 |  EMDBヘッダ EMDBヘッダ |
| FSC (解像度算出) |  emd_27663_fsc.xml emd_27663_fsc.xml | 12.6 KB | 表示 |  FSCデータファイル FSCデータファイル |
| 画像 |  emd_27663.png emd_27663.png | 180.3 KB | ||
| Filedesc metadata |  emd-27663.cif.gz emd-27663.cif.gz | 8.2 KB | ||
| その他 |  emd_27663_additional_1.map.gz emd_27663_additional_1.map.gz emd_27663_half_map_1.map.gz emd_27663_half_map_1.map.gz emd_27663_half_map_2.map.gz emd_27663_half_map_2.map.gz | 677 MB 200.2 MB 200.2 MB | ||
| アーカイブディレクトリ |  http://ftp.pdbj.org/pub/emdb/structures/EMD-27663 http://ftp.pdbj.org/pub/emdb/structures/EMD-27663 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27663 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27663 | HTTPS FTP |
-検証レポート
| 文書・要旨 |  emd_27663_validation.pdf.gz emd_27663_validation.pdf.gz | 845.6 KB | 表示 |  EMDB検証レポート EMDB検証レポート |
|---|---|---|---|---|
| 文書・詳細版 |  emd_27663_full_validation.pdf.gz emd_27663_full_validation.pdf.gz | 845.2 KB | 表示 | |
| XML形式データ |  emd_27663_validation.xml.gz emd_27663_validation.xml.gz | 21.6 KB | 表示 | |
| CIF形式データ |  emd_27663_validation.cif.gz emd_27663_validation.cif.gz | 28 KB | 表示 | |
| アーカイブディレクトリ |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-27663 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-27663 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-27663 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-27663 | HTTPS FTP |
-関連構造データ
| 関連構造データ |  8dqxMC  8dqwC  8dqzC  8dr0C  8dr1C  8dr3C  8dr4C  8dr5C  8dr6C  8dr7C C: 同じ文献を引用 ( M: このマップから作成された原子モデル |
|---|---|
| 類似構造データ | 類似検索 - 機能・相同性  F&H 検索 F&H 検索 |
- リンク
リンク
| EMDBのページ |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| 「今月の分子」の関連する項目 |
- マップ
マップ
| ファイル |  ダウンロード / ファイル: emd_27663.map.gz / 形式: CCP4 / 大きさ: 216 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) ダウンロード / ファイル: emd_27663.map.gz / 形式: CCP4 / 大きさ: 216 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 注釈 | Open RFC-PCNA | ||||||||||||||||||||||||||||||||||||
| 投影像・断面図 | 画像のコントロール
画像は Spider により作成 | ||||||||||||||||||||||||||||||||||||
| ボクセルのサイズ | X=Y=Z: 0.826 Å | ||||||||||||||||||||||||||||||||||||
| 密度 |
| ||||||||||||||||||||||||||||||||||||
| 対称性 | 空間群: 1 | ||||||||||||||||||||||||||||||||||||
| 詳細 | EMDB XML:
|
-添付データ
-追加マップ: Density modified and 1.5x resampled
| ファイル | emd_27663_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 注釈 | Density modified and 1.5x resampled | ||||||||||||
| 投影像・断面図 |
| ||||||||||||
| 密度ヒストグラム |
-ハーフマップ: A
| ファイル | emd_27663_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 注釈 | A | ||||||||||||
| 投影像・断面図 |
| ||||||||||||
| 密度ヒストグラム |
-ハーフマップ: B
| ファイル | emd_27663_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 注釈 | B | ||||||||||||
| 投影像・断面図 |
| ||||||||||||
| 密度ヒストグラム |
- 試料の構成要素
試料の構成要素
+全体 : Open state of RFC:PCNA bound to a 3' ss/dsDNA junction
+超分子 #1: Open state of RFC:PCNA bound to a 3' ss/dsDNA junction
+分子 #1: Replication factor C subunit 1
+分子 #2: Replication factor C subunit 4
+分子 #3: Replication factor C subunit 3
+分子 #4: Replication factor C subunit 2
+分子 #5: Replication factor C subunit 5
+分子 #6: Proliferating cell nuclear antigen
+分子 #7: DNA (5'-D(*TP*TP*TP*TP*TP*T)-3')
+分子 #8: DNA (5'-D(P*TP*CP*CP*GP*AP*GP*CP*GP*AP*A)-3')
+分子 #9: DNA (5'-D(P*TP*TP*TP*GP*CP*CP*CP*GP*GP*A)-3')
+分子 #10: PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER
+分子 #11: MAGNESIUM ION
+分子 #12: GUANOSINE-5'-DIPHOSPHATE
+分子 #13: water
-実験情報
-構造解析
| 手法 | クライオ電子顕微鏡法 |
|---|---|
 解析 解析 | 単粒子再構成法 |
| 試料の集合状態 | particle |
- 試料調製
試料調製
| 濃度 | 1.0 mg/mL | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 緩衝液 | pH: 7.6 構成要素:
| ||||||||
| グリッド | モデル: Quantifoil / 材質: GOLD / メッシュ: 400 / 支持フィルム - 材質: GRAPHENE OXIDE / 支持フィルム - トポロジー: CONTINUOUS | ||||||||
| 凍結 | 凍結剤: ETHANE / チャンバー内湿度: 100 % / チャンバー内温度: 297 K / 装置: FEI VITROBOT MARK IV |
- 電子顕微鏡法
電子顕微鏡法
| 顕微鏡 | FEI TITAN KRIOS |
|---|---|
| 撮影 | フィルム・検出器のモデル: GATAN K3 (6k x 4k) / 平均電子線量: 66.0 e/Å2 |
| 電子線 | 加速電圧: 300 kV / 電子線源:  FIELD EMISSION GUN FIELD EMISSION GUN |
| 電子光学系 | 照射モード: FLOOD BEAM / 撮影モード: BRIGHT FIELD / 最大 デフォーカス(公称値): 2.0 µm 最小 デフォーカス(公称値): 0.7000000000000001 µm |
| 実験機器 |  モデル: Titan Krios / 画像提供: FEI Company |
 ムービー
ムービー コントローラー
コントローラー




















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)