[English] 日本語
 Yorodumi
Yorodumi- EMDB-2751: Structure of the ryanodine receptor at resolution of 6.1 A in clo... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-2751 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of the ryanodine receptor at resolution of 6.1 A in closed state | |||||||||
 Map data Map data | reconstruction of rye reconstituted in lipid nanodiscs in closed state | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | calcium binding / ion channel / muscular contraction / conformational changes. | |||||||||
| Function / homology |  Function and homology information Function and homology informationATP-gated ion channel activity / terminal cisterna / ryanodine-sensitive calcium-release channel activity / ryanodine receptor complex / release of sequestered calcium ion into cytosol by sarcoplasmic reticulum / ossification involved in bone maturation / cellular response to caffeine / skin development / organelle membrane / intracellularly gated calcium channel activity ...ATP-gated ion channel activity / terminal cisterna / ryanodine-sensitive calcium-release channel activity / ryanodine receptor complex / release of sequestered calcium ion into cytosol by sarcoplasmic reticulum / ossification involved in bone maturation / cellular response to caffeine / skin development / organelle membrane / intracellularly gated calcium channel activity / smooth endoplasmic reticulum / outflow tract morphogenesis / toxic substance binding / striated muscle contraction / voltage-gated calcium channel activity / skeletal muscle fiber development / release of sequestered calcium ion into cytosol / sarcoplasmic reticulum membrane / cellular response to calcium ion / muscle contraction / sarcoplasmic reticulum / sarcolemma / calcium ion transmembrane transport / Z disc / calcium channel activity / intracellular calcium ion homeostasis / disordered domain specific binding / protein homotetramerization / transmembrane transporter binding / calmodulin binding / calcium ion binding / ATP binding / identical protein binding / membrane Similarity search - Function | |||||||||
| Biological species |  | |||||||||
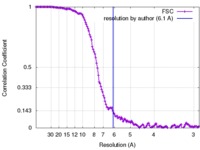
| Method | single particle reconstruction / cryo EM / Resolution: 6.1 Å | |||||||||
 Authors Authors | Efremov RG / Leitner A / Aebersold R / Raunser S | |||||||||
 Citation Citation |  Journal: Nature / Year: 2015 Journal: Nature / Year: 2015Title: Architecture and conformational switch mechanism of the ryanodine receptor. Authors: Rouslan G Efremov / Alexander Leitner / Ruedi Aebersold / Stefan Raunser /    Abstract: Muscle contraction is initiated by the release of calcium (Ca(2+)) from the sarcoplasmic reticulum into the cytoplasm of myocytes through ryanodine receptors (RyRs). RyRs are homotetrameric channels ...Muscle contraction is initiated by the release of calcium (Ca(2+)) from the sarcoplasmic reticulum into the cytoplasm of myocytes through ryanodine receptors (RyRs). RyRs are homotetrameric channels with a molecular mass of more than 2.2 megadaltons that are regulated by several factors, including ions, small molecules and proteins. Numerous mutations in RyRs have been associated with human diseases. The molecular mechanism underlying the complex regulation of RyRs is poorly understood. Using electron cryomicroscopy, here we determine the architecture of rabbit RyR1 at a resolution of 6.1 Å. We show that the cytoplasmic moiety of RyR1 contains two large α-solenoid domains and several smaller domains, with folds suggestive of participation in protein-protein interactions. The transmembrane domain represents a chimaera of voltage-gated sodium and pH-activated ion channels. We identify the calcium-binding EF-hand domain and show that it functions as a conformational switch allosterically gating the channel. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_2751.map.gz emd_2751.map.gz | 18.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-2751-v30.xml emd-2751-v30.xml emd-2751.xml emd-2751.xml | 13.1 KB 13.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_2751_fsc.xml emd_2751_fsc.xml | 12.2 KB | Display |  FSC data file FSC data file |
| Images |  2751-map-snapshot.jpg 2751-map-snapshot.jpg | 67.2 KB | ||
| Masks |  emd_2751_msk_1.map emd_2751_msk_1.map | 216 MB |  Mask map Mask map | |
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-2751 http://ftp.pdbj.org/pub/emdb/structures/EMD-2751 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-2751 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-2751 | HTTPS FTP |
-Related structure data
| Related structure data |  4uwaMC  2752C  4uweC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_2751.map.gz / Format: CCP4 / Size: 210.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_2751.map.gz / Format: CCP4 / Size: 210.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | reconstruction of rye reconstituted in lipid nanodiscs in closed state | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.42 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Segmentation: The mask used for calculating FSC
| Annotation | The mask used for calculating FSC | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| File |  emd_2751_msk_1.map emd_2751_msk_1.map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Ryanodine receptor 1 (calcium release channel) from rabbit
| Entire | Name: Ryanodine receptor 1 (calcium release channel) from rabbit |
|---|---|
| Components |
|
-Supramolecule #1000: Ryanodine receptor 1 (calcium release channel) from rabbit
| Supramolecule | Name: Ryanodine receptor 1 (calcium release channel) from rabbit type: sample / ID: 1000 / Details: protein was reconstituted in lipid nanodiscs / Oligomeric state: tetramer / Number unique components: 1 |
|---|---|
| Molecular weight | Theoretical: 2.26 MDa |
-Macromolecule #1: Ryanodine receptor 1
| Macromolecule | Name: Ryanodine receptor 1 / type: protein_or_peptide / ID: 1 / Name.synonym: Skeletal muscle calcium release channel / Number of copies: 4 / Oligomeric state: tetramer / Recombinant expression: No |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 2.26 MDa |
| Sequence | UniProtKB: Ryanodine receptor 1 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2 mg/mL |
|---|---|
| Buffer | pH: 7.4 Details: 10 mM MOPS, 200 mM NaCl, 1mM EGTA, 0.2% fluorinated octyl-maltoside |
| Grid | Details: Quantifoil R 2/1 holey carbon grid |
| Vitrification | Cryogen name: ETHANE / Instrument: HOMEMADE PLUNGER Method: protein solution was applied on glow discharged grid and blotted for 4 seconds before plunging |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Cs | 0 |
| Temperature | Min: 80 K |
| Date | Feb 3, 2014 |
| Image recording | Category: CCD / Film or detector model: FEI FALCON II (4k x 4k) / Number real images: 2320 / Average electron dose: 20 e/Å2 Details: Images were collected automatically with CPU software and using movie mode with 7 frames per image. |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 0.003 µm / Nominal defocus min: 0.001 µm / Nominal magnification: 47000 / Cs: mm |
| Sample stage | Specimen holder: liquid nitorgen cooled / Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
-Atomic model buiding 1
| Initial model | PDB ID: |
|---|---|
| Software | Name:  Chimera Chimera |
| Details | The domains with known crystal structures: 2AOX, 4ERT, 2R9R, or modelled with structure prediction programs were separately fitted in Chimera. The remaining domains were build ab initio in coot |
| Refinement | Space: REAL / Protocol: RIGID BODY FIT |
| Output model |  PDB-4uwa: |
-Atomic model buiding 2
| Initial model | PDB ID: |
|---|---|
| Software | Name:  Chimera Chimera |
| Details | The domains with known crystal structures: 2AOX, 4ERT, 2R9R, or modelled with structure prediction programs were separately fitted in Chimera. The remaining domains were build ab initio in coot |
| Refinement | Space: REAL / Protocol: RIGID BODY FIT |
| Output model |  PDB-4uwa: |
-Atomic model buiding 3
| Initial model | PDB ID: |
|---|---|
| Software | Name:  Chimera Chimera |
| Details | The domains with known crystal structures: 2AOX, 4ERT, 2R9R, or modelled with structure prediction programs were separately fitted in Chimera. The remaining domains were build ab initio in coot |
| Refinement | Space: REAL / Protocol: RIGID BODY FIT |
| Output model |  PDB-4uwa: |
 Movie
Movie Controller
Controller














 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)