+ データを開く
データを開く
- 基本情報
基本情報
| 登録情報 |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| タイトル | Saccharomyces cerevisiae Ufd1/Npl4/Cdc48 complex bound to two ubiquitin moieties and one unfolded ubiquitin in presence of SUMO-ubiquitin(K48polyUb)-mEOS and ATP, state 2 (uC) | |||||||||
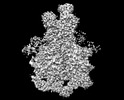
 マップデータ マップデータ | composite map of the ubiquitin unfolded state 'uC' | |||||||||
 試料 試料 |
| |||||||||
 キーワード キーワード | ATPASE / ATPASE COMPLEX / UBIQUITIN / SUMO / SMT3 / QUALITY CONTROL / MOTOR PROTEIN | |||||||||
| 機能・相同性 |  機能・相同性情報 機能・相同性情報: / RAS processing / Regulation of PTEN localization / : / UCH proteinases / : / Aggrephagy / Pexophagy / PINK1-PRKN Mediated Mitophagy / : ...: / RAS processing / Regulation of PTEN localization / : / UCH proteinases / : / Aggrephagy / Pexophagy / PINK1-PRKN Mediated Mitophagy / : / SCF complex disassembly in response to cadmium stress / mitotic DNA replication termination / Ovarian tumor domain proteases / Cdc48p-Npl4p-Vms1p AAA ATPase complex / Doa10p ubiquitin ligase complex / stress-induced homeostatically regulated protein degradation pathway / Peroxisomal protein import / : / sister chromatid biorientation / protein localization to vacuole / endoplasmic reticulum membrane fusion / ribophagy / Hrd1p ubiquitin ligase ERAD-L complex / DNA replication termination / RQC complex / : / Endosomal Sorting Complex Required For Transport (ESCRT) / mitochondria-associated ubiquitin-dependent protein catabolic process / positive regulation of mitochondrial fusion / cytoplasm protein quality control by the ubiquitin-proteasome system / HSF1 activation / nuclear protein quality control by the ubiquitin-proteasome system / protein-containing complex disassembly / : / protein transport to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway / endosome to plasma membrane protein transport / protein phosphatase regulator activity / Translesion synthesis by REV1 / : / : / : / piecemeal microautophagy of the nucleus / : / mating projection tip / Recruitment and ATM-mediated phosphorylation of repair and signaling proteins at DNA double strand breaks / : / mitotic spindle disassembly / Protein methylation / VCP-NPL4-UFD1 AAA ATPase complex / replisome / ribosome-associated ubiquitin-dependent protein catabolic process / vesicle-fusing ATPase / K48-linked polyubiquitin modification-dependent protein binding / nuclear outer membrane-endoplasmic reticulum membrane network / retrograde protein transport, ER to cytosol / nonfunctional rRNA decay / Ubiquitin-Mediated Degradation of Phosphorylated Cdc25A / Regulation of PTEN stability and activity / CDK-mediated phosphorylation and removal of Cdc6 / FBXL7 down-regulates AURKA during mitotic entry and in early mitosis / KEAP1-NFE2L2 pathway / Neddylation / Formation of TC-NER Pre-Incision Complex / Orc1 removal from chromatin / MAPK6/MAPK4 signaling / SRP-dependent cotranslational protein targeting to membrane / GTP hydrolysis and joining of the 60S ribosomal subunit / Gap-filling DNA repair synthesis and ligation in TC-NER / protein quality control for misfolded or incompletely synthesized proteins / Formation of a pool of free 40S subunits / Nonsense Mediated Decay (NMD) independent of the Exon Junction Complex (EJC) / Nonsense Mediated Decay (NMD) enhanced by the Exon Junction Complex (EJC) / Antigen processing: Ubiquitination & Proteasome degradation / L13a-mediated translational silencing of Ceruloplasmin expression / Dual incision in TC-NER / polyubiquitin modification-dependent protein binding / autophagosome maturation / mRNA transport / Ub-specific processing proteases / ATP metabolic process / ERAD pathway / Neutrophil degranulation / rescue of stalled cytosolic ribosome / ubiquitin binding / macroautophagy / modification-dependent protein catabolic process / positive regulation of protein localization to nucleus / protein tag activity / peroxisome / ubiquitin-dependent protein catabolic process / nuclear membrane / proteasome-mediated ubiquitin-dependent protein catabolic process / protein ubiquitination / ubiquitin protein ligase binding / endoplasmic reticulum membrane / perinuclear region of cytoplasm / ATP hydrolysis activity / mitochondrion / ATP binding / identical protein binding 類似検索 - 分子機能 | |||||||||
| 生物種 |  | |||||||||
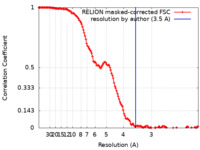
| 手法 | 単粒子再構成法 / クライオ電子顕微鏡法 / 解像度: 3.5 Å | |||||||||
 データ登録者 データ登録者 | Lee HG / Lima CD | |||||||||
| 資金援助 |  米国, 2件 米国, 2件
| |||||||||
 引用 引用 |  ジャーナル: Proc Natl Acad Sci U S A / 年: 2023 ジャーナル: Proc Natl Acad Sci U S A / 年: 2023タイトル: SUMO enhances unfolding of SUMO-polyubiquitin-modified substrates by the Ufd1/Npl4/Cdc48 complex. 著者: Hyein G Lee / Abigail A Lemmon / Christopher D Lima /  要旨: The Ufd1/Npl4/Cdc48 complex is a universal protein segregase that plays key roles in eukaryotic cellular processes. Its functions orchestrating the clearance or removal of polyubiquitylated targets ...The Ufd1/Npl4/Cdc48 complex is a universal protein segregase that plays key roles in eukaryotic cellular processes. Its functions orchestrating the clearance or removal of polyubiquitylated targets are established; however, prior studies suggest that the complex also targets substrates modified by the ubiquitin-like protein SUMO. Here, we show that interactions between Ufd1 and SUMO enhance unfolding of substrates modified by SUMO-polyubiquitin hybrid chains by the budding yeast Ufd1/Npl4/Cdc48 complex compared to substrates modified by polyubiquitin chains, a difference that is accentuated when the complex has a choice between these substrates. Incubating Ufd1/Npl4/Cdc48 with a substrate modified by a SUMO-polyubiquitin hybrid chain produced a series of single-particle cryo-EM structures that reveal features of interactions between Ufd1/Npl4/Cdc48 and ubiquitin prior to and during unfolding of ubiquitin. These results are consistent with cellular functions for SUMO and ubiquitin modifications and support a physical model wherein Ufd1/Npl4/Cdc48, SUMO, and ubiquitin conjugation pathways converge to promote clearance of proteins modified with SUMO and polyubiquitin. | |||||||||
| 履歴 |
|
- 構造の表示
構造の表示
| 添付画像 |
|---|
- ダウンロードとリンク
ダウンロードとリンク
-EMDBアーカイブ
| マップデータ |  emd_27277.map.gz emd_27277.map.gz | 25.4 MB |  EMDBマップデータ形式 EMDBマップデータ形式 | |
|---|---|---|---|---|
| ヘッダ (付随情報) |  emd-27277-v30.xml emd-27277-v30.xml emd-27277.xml emd-27277.xml | 54 KB 54 KB | 表示 表示 |  EMDBヘッダ EMDBヘッダ |
| FSC (解像度算出) |  emd_27277_fsc.xml emd_27277_fsc.xml | 13.7 KB | 表示 |  FSCデータファイル FSCデータファイル |
| 画像 |  emd_27277.png emd_27277.png | 92.1 KB | ||
| Filedesc metadata |  emd-27277.cif.gz emd-27277.cif.gz | 8.3 KB | ||
| その他 |  emd_27277_additional_1.map.gz emd_27277_additional_1.map.gz emd_27277_additional_10.map.gz emd_27277_additional_10.map.gz emd_27277_additional_11.map.gz emd_27277_additional_11.map.gz emd_27277_additional_12.map.gz emd_27277_additional_12.map.gz emd_27277_additional_13.map.gz emd_27277_additional_13.map.gz emd_27277_additional_2.map.gz emd_27277_additional_2.map.gz emd_27277_additional_3.map.gz emd_27277_additional_3.map.gz emd_27277_additional_4.map.gz emd_27277_additional_4.map.gz emd_27277_additional_5.map.gz emd_27277_additional_5.map.gz emd_27277_additional_6.map.gz emd_27277_additional_6.map.gz emd_27277_additional_7.map.gz emd_27277_additional_7.map.gz emd_27277_additional_8.map.gz emd_27277_additional_8.map.gz emd_27277_additional_9.map.gz emd_27277_additional_9.map.gz emd_27277_half_map_1.map.gz emd_27277_half_map_1.map.gz emd_27277_half_map_2.map.gz emd_27277_half_map_2.map.gz | 17.7 MB 171.4 MB 171.2 MB 171.1 MB 171.4 MB 171.1 MB 12.8 MB 171.2 MB 12.4 MB 8.6 MB 10.1 MB 171.3 MB 171.3 MB 171.4 MB 171.2 MB | ||
| アーカイブディレクトリ |  http://ftp.pdbj.org/pub/emdb/structures/EMD-27277 http://ftp.pdbj.org/pub/emdb/structures/EMD-27277 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27277 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27277 | HTTPS FTP |
-関連構造データ
- リンク
リンク
| EMDBのページ |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| 「今月の分子」の関連する項目 |
- マップ
マップ
| ファイル |  ダウンロード / ファイル: emd_27277.map.gz / 形式: CCP4 / 大きさ: 216 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) ダウンロード / ファイル: emd_27277.map.gz / 形式: CCP4 / 大きさ: 216 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 注釈 | composite map of the ubiquitin unfolded state 'uC' | ||||||||||||||||||||||||||||||||||||
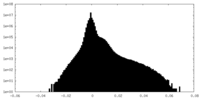
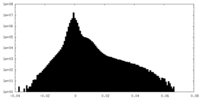
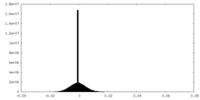
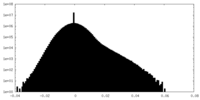
| 投影像・断面図 | 画像のコントロール
画像は Spider により作成 | ||||||||||||||||||||||||||||||||||||
| ボクセルのサイズ | X=Y=Z: 1.064 Å | ||||||||||||||||||||||||||||||||||||
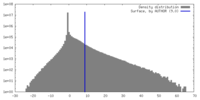
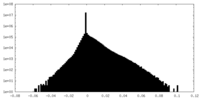
| 密度 |
| ||||||||||||||||||||||||||||||||||||
| 対称性 | 空間群: 1 | ||||||||||||||||||||||||||||||||||||
| 詳細 | EMDB XML:
|
-添付データ
+追加マップ: post-process overall refinement map of the ubiquitin unfolded...
+追加マップ: focused refinement half map 2 of the Cdc48...
+追加マップ: focused refinement half map 1 of the D1/D2...
+追加マップ: focused refinement half map 1 of the upper...
+追加マップ: focused refinement half map 1 of the Cdc48...
+追加マップ: focused refinement half map 2 of the upper...
+追加マップ: post-processed focused refinement map of the Cdc48 hexamer...
+追加マップ: focused refinement half map 2 of the D1/D2...
+追加マップ: post-processed focused refinement map of the D1/D2 domains...
+追加マップ: post-processed focused refinement map of the upper Ufd1/Npl4/polyubiquitin...
+追加マップ: post-processed focused refinement map of the Ufd1/Npl4/polyubiquitin density...
+追加マップ: focused refinement half map 1 of the Ufd1/Npl4/polyubiquitin...
+追加マップ: focused refinement half map 2 of the Ufd1/Npl4/polyubiquitin...
+ハーフマップ: overall refinement half map 2 of the ubiquitin unfolded state 'uC'
+ハーフマップ: overall refinement half map 1 of the ubiquitin unfolded state 'uC'
- 試料の構成要素
試料の構成要素
+全体 : Saccharomyces cerevisiae Ufd1/Npl4/Cdc48 complex bound to two fol...
+超分子 #1: Saccharomyces cerevisiae Ufd1/Npl4/Cdc48 complex bound to two fol...
+超分子 #2: Saccharomyces cerevisiae Ufd1/Npl4/Cdc48 complex bound to two fol...
+超分子 #3: Cdc48 hexamer
+超分子 #4: Two folded ubiqutin moieties and one unfolded ubiquitin bound to ...
+超分子 #5: D1/D2 ATPase domains of the Cdc48 hexamer
+超分子 #6: Two folded ubiqutin moieties bound to Ufd1/Npl4
+分子 #1: Cell division control protein 48
+分子 #2: Nuclear protein localization protein 4
+分子 #3: Ubiquitin fusion degradation protein 1
+分子 #4: Ubiquitin
+分子 #5: ADENOSINE-5'-TRIPHOSPHATE
+分子 #6: ADENOSINE-5'-DIPHOSPHATE
+分子 #7: ZINC ION
-実験情報
-構造解析
| 手法 | クライオ電子顕微鏡法 |
|---|---|
 解析 解析 | 単粒子再構成法 |
| 試料の集合状態 | particle |
- 試料調製
試料調製
| 濃度 | 2 mg/mL |
|---|---|
| 緩衝液 | pH: 8 詳細: 20 mM HEPES pH 8.0, 150 mM NaCl, 0.1 mM TCEP, 1 mM MgCl2, 5 mM ATP. Added 0.05% CHAPSO before vitrification. |
| 凍結 | 凍結剤: ETHANE / チャンバー内湿度: 100 % / チャンバー内温度: 295 K / 装置: FEI VITROBOT MARK IV / 詳細: 8 s wait, 4 s blot before plunging. |
| 詳細 | Ufd1/Npl4/Cdc48 was pre-incubated with SUMO-ubiquitin(K48polyUb)-mEOS and ATP |
- 電子顕微鏡法
電子顕微鏡法
| 顕微鏡 | FEI TITAN KRIOS |
|---|---|
| 撮影 | フィルム・検出器のモデル: GATAN K3 (6k x 4k) / 平均電子線量: 70.577 e/Å2 |
| 電子線 | 加速電圧: 300 kV / 電子線源:  FIELD EMISSION GUN FIELD EMISSION GUN |
| 電子光学系 | 照射モード: FLOOD BEAM / 撮影モード: BRIGHT FIELD 最大 デフォーカス(公称値): 2.8000000000000003 µm 最小 デフォーカス(公称値): 1.2 µm |
| 実験機器 |  モデル: Titan Krios / 画像提供: FEI Company |
 ムービー
ムービー コントローラー
コントローラー


































 X (Sec.)
X (Sec.) Y (Row.)
Y (Row.) Z (Col.)
Z (Col.)