[English] 日本語
 Yorodumi
Yorodumi- EMDB-27273: Saccharomyces cerevisiae Ufd1/Npl4/Cdc48 complex unbound but in t... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Saccharomyces cerevisiae Ufd1/Npl4/Cdc48 complex unbound but in the presence of SUMO-ubiquitin(K48polyUb)-mEOS and ATP | |||||||||
 Map data Map data | composite map of substrate unbound class of particles. This class represents the portion of particles where Ufd1/Npl4/Cdc48 was not bound to the SUMO-ubiquitin(K48polyUb)-mEOS substrate. Composite map represents the composite map of overall and focused refinements under this deposition. | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | ATPASE / ATPASE COMPLEX / UBIQUITIN / SUMO / SMT3 / QUALITY CONTROL / MOTOR PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationSCF complex disassembly in response to cadmium stress / mitotic DNA replication termination / Ovarian tumor domain proteases / Cdc48p-Npl4p-Vms1p AAA ATPase complex / Doa10p ubiquitin ligase complex / stress-induced homeostatically regulated protein degradation pathway / sister chromatid biorientation / protein localization to vacuole / endoplasmic reticulum membrane fusion / ribophagy ...SCF complex disassembly in response to cadmium stress / mitotic DNA replication termination / Ovarian tumor domain proteases / Cdc48p-Npl4p-Vms1p AAA ATPase complex / Doa10p ubiquitin ligase complex / stress-induced homeostatically regulated protein degradation pathway / sister chromatid biorientation / protein localization to vacuole / endoplasmic reticulum membrane fusion / ribophagy / Hrd1p ubiquitin ligase ERAD-L complex / DNA replication termination / RQC complex / mitochondria-associated ubiquitin-dependent protein catabolic process / positive regulation of mitochondrial fusion / cytoplasm protein quality control by the ubiquitin-proteasome system / HSF1 activation / nuclear protein quality control by the ubiquitin-proteasome system / protein-containing complex disassembly / protein transport to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway / endosome to plasma membrane protein transport / protein phosphatase regulator activity / : / piecemeal microautophagy of the nucleus / mating projection tip / mitotic spindle disassembly / Protein methylation / VCP-NPL4-UFD1 AAA ATPase complex / replisome / ribosome-associated ubiquitin-dependent protein catabolic process / vesicle-fusing ATPase / K48-linked polyubiquitin modification-dependent protein binding / nuclear outer membrane-endoplasmic reticulum membrane network / retrograde protein transport, ER to cytosol / nonfunctional rRNA decay / KEAP1-NFE2L2 pathway / Neddylation / protein quality control for misfolded or incompletely synthesized proteins / polyubiquitin modification-dependent protein binding / autophagosome maturation / mRNA transport / ATP metabolic process / ERAD pathway / Neutrophil degranulation / rescue of stalled cytosolic ribosome / ubiquitin binding / macroautophagy / positive regulation of protein localization to nucleus / ubiquitin-dependent protein catabolic process / nuclear membrane / proteasome-mediated ubiquitin-dependent protein catabolic process / ubiquitin protein ligase binding / endoplasmic reticulum membrane / perinuclear region of cytoplasm / ATP hydrolysis activity / mitochondrion / ATP binding / identical protein binding / nucleus / cytosol / cytoplasm Similarity search - Function | |||||||||
| Biological species |  | |||||||||
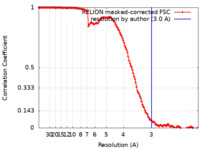
| Method | single particle reconstruction / cryo EM / Resolution: 3.0 Å | |||||||||
 Authors Authors | Lee HG / Lima CD | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2023 Journal: Proc Natl Acad Sci U S A / Year: 2023Title: SUMO enhances unfolding of SUMO-polyubiquitin-modified substrates by the Ufd1/Npl4/Cdc48 complex. Authors: Hyein G Lee / Abigail A Lemmon / Christopher D Lima /  Abstract: The Ufd1/Npl4/Cdc48 complex is a universal protein segregase that plays key roles in eukaryotic cellular processes. Its functions orchestrating the clearance or removal of polyubiquitylated targets ...The Ufd1/Npl4/Cdc48 complex is a universal protein segregase that plays key roles in eukaryotic cellular processes. Its functions orchestrating the clearance or removal of polyubiquitylated targets are established; however, prior studies suggest that the complex also targets substrates modified by the ubiquitin-like protein SUMO. Here, we show that interactions between Ufd1 and SUMO enhance unfolding of substrates modified by SUMO-polyubiquitin hybrid chains by the budding yeast Ufd1/Npl4/Cdc48 complex compared to substrates modified by polyubiquitin chains, a difference that is accentuated when the complex has a choice between these substrates. Incubating Ufd1/Npl4/Cdc48 with a substrate modified by a SUMO-polyubiquitin hybrid chain produced a series of single-particle cryo-EM structures that reveal features of interactions between Ufd1/Npl4/Cdc48 and ubiquitin prior to and during unfolding of ubiquitin. These results are consistent with cellular functions for SUMO and ubiquitin modifications and support a physical model wherein Ufd1/Npl4/Cdc48, SUMO, and ubiquitin conjugation pathways converge to promote clearance of proteins modified with SUMO and polyubiquitin. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_27273.map.gz emd_27273.map.gz | 25.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-27273-v30.xml emd-27273-v30.xml emd-27273.xml emd-27273.xml | 42.4 KB 42.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_27273_fsc.xml emd_27273_fsc.xml | 13.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_27273.png emd_27273.png | 81.8 KB | ||
| Filedesc metadata |  emd-27273.cif.gz emd-27273.cif.gz | 7.7 KB | ||
| Others |  emd_27273_additional_1.map.gz emd_27273_additional_1.map.gz emd_27273_additional_10.map.gz emd_27273_additional_10.map.gz emd_27273_additional_2.map.gz emd_27273_additional_2.map.gz emd_27273_additional_3.map.gz emd_27273_additional_3.map.gz emd_27273_additional_4.map.gz emd_27273_additional_4.map.gz emd_27273_additional_5.map.gz emd_27273_additional_5.map.gz emd_27273_additional_6.map.gz emd_27273_additional_6.map.gz emd_27273_additional_7.map.gz emd_27273_additional_7.map.gz emd_27273_additional_8.map.gz emd_27273_additional_8.map.gz emd_27273_additional_9.map.gz emd_27273_additional_9.map.gz emd_27273_half_map_1.map.gz emd_27273_half_map_1.map.gz emd_27273_half_map_2.map.gz emd_27273_half_map_2.map.gz | 13.3 MB 171.4 MB 14.7 MB 18.5 MB 10.6 MB 171.5 MB 171.5 MB 171.6 MB 171.4 MB 171.7 MB 171.4 MB 171.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-27273 http://ftp.pdbj.org/pub/emdb/structures/EMD-27273 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27273 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27273 | HTTPS FTP |
-Related structure data
| Related structure data |  8darMC  8dasC  8datC  8dauC  8davC  8dawC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_27273.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_27273.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | composite map of substrate unbound class of particles. This class represents the portion of particles where Ufd1/Npl4/Cdc48 was not bound to the SUMO-ubiquitin(K48polyUb)-mEOS substrate. Composite map represents the composite map of overall and focused refinements under this deposition. | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.064 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
+Additional map: focused refinement of the D1/D2 domains of Cdc48...
+Additional map: focused refinement half map 1 of the D1/D2...
+Additional map: focused refinement of cdc48 of substrate unbound class...
+Additional map: overall refinement of substrate unbound class of particles,...
+Additional map: focused refinement of the central densities of Ufd1/Npl4...
+Additional map: focused refinement half map 2 of the D1/D2...
+Additional map: focused refinement half map 1 of Cdc48 of...
+Additional map: focused refinement half map 1 of the central...
+Additional map: focused refinement half map 2 of Cdc48 of...
+Additional map: focused refinement half map 2 of the central...
+Half map: overall refinement half map 2 of substrate unbound class of particles
+Half map: overall refinement half map 1 of substrate unbound class of particles
- Sample components
Sample components
-Entire : Saccharomyces cerevisiae Ufd1/Npl4/Cdc48 complex unbound but in t...
| Entire | Name: Saccharomyces cerevisiae Ufd1/Npl4/Cdc48 complex unbound but in the presence of SUMO-ubiquitin(K48polyUb)-mEOS and ATP |
|---|---|
| Components |
|
-Supramolecule #1: Saccharomyces cerevisiae Ufd1/Npl4/Cdc48 complex unbound but in t...
| Supramolecule | Name: Saccharomyces cerevisiae Ufd1/Npl4/Cdc48 complex unbound but in the presence of SUMO-ubiquitin(K48polyUb)-mEOS and ATP type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 Details: This class represents the portion of particles where Ufd1/Npl4/Cdc48 was not bound to the SUMO-ubiquitin(K48polyUb)-mEOS substrate. Composite map represents the composite map of overall and ...Details: This class represents the portion of particles where Ufd1/Npl4/Cdc48 was not bound to the SUMO-ubiquitin(K48polyUb)-mEOS substrate. Composite map represents the composite map of overall and focused refinements under this deposition. |
|---|
-Supramolecule #2: Cdc48 hexamer
| Supramolecule | Name: Cdc48 hexamer / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
-Supramolecule #3: Ufd1/Npl4 tower
| Supramolecule | Name: Ufd1/Npl4 tower / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #2-#3 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Cell division control protein 48
| Macromolecule | Name: Cell division control protein 48 / type: protein_or_peptide / ID: 1 / Number of copies: 6 / Enantiomer: LEVO / EC number: vesicle-fusing ATPase |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 92.389195 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: GSHMGEEHKP LLDASGVDPR EEDKTATAIL RRKKKDNMLL VDDAINDDNS VIAINSNTMD KLELFRGDTV LVKGKKRKDT VLIVLIDDE LEDGACRINR VVRNNLRIRL GDLVTIHPCP DIKYATRISV LPIADTIEGI TGNLFDVFLK PYFVEAYRPV R KGDHFVVR ...String: GSHMGEEHKP LLDASGVDPR EEDKTATAIL RRKKKDNMLL VDDAINDDNS VIAINSNTMD KLELFRGDTV LVKGKKRKDT VLIVLIDDE LEDGACRINR VVRNNLRIRL GDLVTIHPCP DIKYATRISV LPIADTIEGI TGNLFDVFLK PYFVEAYRPV R KGDHFVVR GGMRQVEFKV VDVEPEEYAV VAQDTIIHWE GEPINREDEE NNMNEVGYDD IGGCRKQMAQ IREMVELPLR HP QLFKAIG IKPPRGVLMY GPPGTGKTLM ARAVANETGA FFFLINGPEV MSKMAGESES NLRKAFEEAE KNAPAIIFID EID SIAPKR DKTNGEVERR VVSQLLTLMD GMKARSNVVV IAATNRPNSI DPALRRFGRF DREVDIGIPD ATGRLEVLRI HTKN MKLAD DVDLEALAAE THGYVGADIA SLCSEAAMQQ IREKMDLIDL DEDEIDAEVL DSLGVTMDNF RFALGNSNPS ALRET VVES VNVTWDDVGG LDEIKEELKE TVEYPVLHPD QYTKFGLSPS KGVLFYGPPG TGKTLLAKAV ATEVSANFIS VKGPEL LSM WYGESESNIR DIFDKARAAA PTVVFLDELD SIAKARGGSL GDAGGASDRV VNQLLTEMDG MNAKKNVFVI GATNRPD QI DPAILRPGRL DQLIYVPLPD ENARLSILNA QLRKTPLEPG LELTAIAKAT QGFSGADLLY IVQRAAKYAI KDSIEAHR Q HEAEKEVKVE GEDVEMTDEG AKAEQEPEVD PVPYITKEHF AEAMKTAKRS VSDAELRRYE AYSQQMKASR GQFSNFNFN DAPLGTTATD NANSNNSAPS GAGAAFGSNA EEDDDLYS UniProtKB: Cell division control protein 48 |
-Macromolecule #2: Nuclear protein localization protein 4
| Macromolecule | Name: Nuclear protein localization protein 4 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 66.144336 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: GSHMLIRFRS KNGTHRVSCQ ENDLFGTVIE KLVGNLDPNA DVDTFTVCEK PGQGIHAVSE LADRTVMDLG LKHGDMLILN YSDKPANEK DGVNVEIGSV GIDSKGIRQH RYGPLRIKEL AVDEELEKED GLIPRQKSKL CKHGDRGMCE YCSPLPPWDK E YHEKNKIK ...String: GSHMLIRFRS KNGTHRVSCQ ENDLFGTVIE KLVGNLDPNA DVDTFTVCEK PGQGIHAVSE LADRTVMDLG LKHGDMLILN YSDKPANEK DGVNVEIGSV GIDSKGIRQH RYGPLRIKEL AVDEELEKED GLIPRQKSKL CKHGDRGMCE YCSPLPPWDK E YHEKNKIK HISFHSYLKK LNENANKKEN GSSYISPLSE PDFRINKRCH NGHEPWPRGI CSKCQPSAIT LQQQEFRMVD HV EFQKSEI INEFIQAWRY TGMQRFGYMY GSYSKYDNTP LGIKAVVEAI YEPPQHDEQD GLTMDVEQVK NEMLQIDRQA QEM GLSRIG LIFTDLSDAG AGDGSVFCKR HKDSFFLSSL EVIMAARHQT RHPNVSKYSE QGFFSSKFVT CVISGNLEGE IDIS SYQVS TEAEALVTAD MISGSTFPSM AYINDTTDER YVPEIFYMKS NEYGITVKEN AKPAFPVDYL LVTLTHGFPN TDTET NSKF VSSTGFPWSN RQAMGQSQDY QELKKYLFNV ASSGDFNLLH EKISNFHLLL YINSLQILSP DEWKLLIESA VKNEWE ESL LKLVSSAGWQ TLVMILQESG UniProtKB: Nuclear protein localization protein 4 |
-Macromolecule #3: Ubiquitin fusion degradation protein 1
| Macromolecule | Name: Ubiquitin fusion degradation protein 1 / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 40.001449 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: GSMFSGFSSF GGGNGFVNMP QTFEEFFRCY PIAMMNDRIR KDDANFGGKI FLPPSALSKL SMLNIRYPML FKLTANETGR VTHGGVLEF IAEEGRVYLP QWMMETLGIQ PGSLLQISST DVPLGQFVKL EPQSVDFLDI SDPKAVLENV LRNFSTLTVD D VIEISYNG ...String: GSMFSGFSSF GGGNGFVNMP QTFEEFFRCY PIAMMNDRIR KDDANFGGKI FLPPSALSKL SMLNIRYPML FKLTANETGR VTHGGVLEF IAEEGRVYLP QWMMETLGIQ PGSLLQISST DVPLGQFVKL EPQSVDFLDI SDPKAVLENV LRNFSTLTVD D VIEISYNG KTFKIKILEV KPESSSKSIC VIETDLVTDF APPVGYVEPD YKALKAQQDK EKKNSFGKGQ VLDPSVLGQG SM STRIDYA GIANSSRNKL SKFVGQGQNI SGKAPKAEPK QDIKDMKITF DGEPAKLDLP EGQLFFGFPM VLPKEDEESA AGS KSSEQN FQGQGISLRK SNKRKTKSDH DSSKSKAPKS PEVIEID UniProtKB: Ubiquitin fusion degradation protein 1 |
-Macromolecule #4: ADENOSINE-5'-TRIPHOSPHATE
| Macromolecule | Name: ADENOSINE-5'-TRIPHOSPHATE / type: ligand / ID: 4 / Number of copies: 6 / Formula: ATP |
|---|---|
| Molecular weight | Theoretical: 507.181 Da |
| Chemical component information |  ChemComp-ATP: |
-Macromolecule #5: ADENOSINE-5'-DIPHOSPHATE
| Macromolecule | Name: ADENOSINE-5'-DIPHOSPHATE / type: ligand / ID: 5 / Number of copies: 6 / Formula: ADP |
|---|---|
| Molecular weight | Theoretical: 427.201 Da |
| Chemical component information |  ChemComp-ADP: |
-Macromolecule #6: ZINC ION
| Macromolecule | Name: ZINC ION / type: ligand / ID: 6 / Number of copies: 2 / Formula: ZN |
|---|---|
| Molecular weight | Theoretical: 65.409 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2 mg/mL |
|---|---|
| Buffer | pH: 8 Details: 20 mM HEPES pH 8.0, 150 mM NaCl, 0.1 mM TCEP, 1 mM MgCl2, 5 mM ATP. Added 0.05% CHAPSO before vitrification. |
| Grid | Model: UltrAuFoil R1.2/1.3 / Material: GOLD / Mesh: 300 / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 295 K / Instrument: FEI VITROBOT MARK IV / Details: 8 s wait, 4 s blot before plunging. |
| Details | Ufd1/Npl4/Cdc48 was pre-incubated with SUMO-ubiquitin(K48polyUb)-mEOS and ATP |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 70.577 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.8000000000000003 µm / Nominal defocus min: 1.2 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller

























 X (Sec.)
X (Sec.) Y (Row.)
Y (Row.) Z (Col.)
Z (Col.)