+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
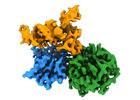
| Title | CryoEM structure of human METTL1-WDR4 in complex with Lys-tRNA | ||||||||||||
 Map data Map data | Human METTL1-WDR4 proteins in complex with Lys tRNA. | ||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | Cancer protein / Methyl transferase / TRANSFERASE | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationinternal mRNA (guanine-N7-)-methyltransferase activity / tRNA (m7G46) methyltransferase complex / tRNA (guanine-N7)-methylation / tRNA stabilization / RNA (guanine-N7)-methylation / tRNA (guanine46-N7)-methyltransferase / tRNA (guanine(46)-N7)-methyltransferase activity / tRNA methyltransferase activator activity / tRNA methyltransferase complex / tRNA modification in the nucleus and cytosol ...internal mRNA (guanine-N7-)-methyltransferase activity / tRNA (m7G46) methyltransferase complex / tRNA (guanine-N7)-methylation / tRNA stabilization / RNA (guanine-N7)-methylation / tRNA (guanine46-N7)-methyltransferase / tRNA (guanine(46)-N7)-methyltransferase activity / tRNA methyltransferase activator activity / tRNA methyltransferase complex / tRNA modification in the nucleus and cytosol / tRNA modification / tRNA methylation / cellular response to stress / Transferases; Transferring one-carbon groups; Methyltransferases / enzyme activator activity / chromosome / tRNA binding / DNA damage response / nucleolus / nucleoplasm / nucleus / cytosol Similarity search - Function | ||||||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.72 Å | ||||||||||||
 Authors Authors | Ruiz-Arroyo VM / Nam Y | ||||||||||||
| Funding support |  United States, 3 items United States, 3 items
| ||||||||||||
 Citation Citation |  Journal: Nature / Year: 2023 Journal: Nature / Year: 2023Title: Structures and mechanisms of tRNA methylation by METTL1-WDR4. Authors: Victor M Ruiz-Arroyo / Rishi Raj / Kesavan Babu / Otgonbileg Onolbaatar / Paul H Roberts / Yunsun Nam /  Abstract: Specific, regulated modification of RNAs is important for proper gene expression. tRNAs are rich with various chemical modifications that affect their stability and function. 7-Methylguanosine (mG) ...Specific, regulated modification of RNAs is important for proper gene expression. tRNAs are rich with various chemical modifications that affect their stability and function. 7-Methylguanosine (mG) at tRNA position 46 is a conserved modification that modulates steady-state tRNA levels to affect cell growth. The METTL1-WDR4 complex generates mG46 in humans, and dysregulation of METTL1-WDR4 has been linked to brain malformation and multiple cancers. Here we show how METTL1 and WDR4 cooperate to recognize RNA substrates and catalyse methylation. A crystal structure of METTL1-WDR4 and cryo-electron microscopy structures of METTL1-WDR4-tRNA show that the composite protein surface recognizes the tRNA elbow through shape complementarity. The cryo-electron microscopy structures of METTL1-WDR4-tRNA with S-adenosylmethionine or S-adenosylhomocysteine along with METTL1 crystal structures provide additional insights into the catalytic mechanism by revealing the active site in multiple states. The METTL1 N terminus couples cofactor binding with conformational changes in the tRNA, the catalytic loop and the WDR4 C terminus, acting as the switch to activate mG methylation. Thus, our structural models explain how post-translational modifications of the METTL1 N terminus can regulate methylation. Together, our work elucidates the core and regulatory mechanisms underlying mG modification by METTL1, providing the framework to understand its contribution to biology and disease. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_27264.map.gz emd_27264.map.gz | 5.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-27264-v30.xml emd-27264-v30.xml emd-27264.xml emd-27264.xml | 19.5 KB 19.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_27264_fsc.xml emd_27264_fsc.xml | 4.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_27264.png emd_27264.png | 80.1 KB | ||
| Masks |  emd_27264_msk_1.map emd_27264_msk_1.map | 11.4 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-27264.cif.gz emd-27264.cif.gz | 6.2 KB | ||
| Others |  emd_27264_additional_1.map.gz emd_27264_additional_1.map.gz emd_27264_half_map_1.map.gz emd_27264_half_map_1.map.gz emd_27264_half_map_2.map.gz emd_27264_half_map_2.map.gz | 10.7 MB 10.6 MB 10.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-27264 http://ftp.pdbj.org/pub/emdb/structures/EMD-27264 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27264 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27264 | HTTPS FTP |
-Validation report
| Summary document |  emd_27264_validation.pdf.gz emd_27264_validation.pdf.gz | 808.4 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_27264_full_validation.pdf.gz emd_27264_full_validation.pdf.gz | 807.9 KB | Display | |
| Data in XML |  emd_27264_validation.xml.gz emd_27264_validation.xml.gz | 11.7 KB | Display | |
| Data in CIF |  emd_27264_validation.cif.gz emd_27264_validation.cif.gz | 14.8 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-27264 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-27264 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-27264 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-27264 | HTTPS FTP |
-Related structure data
| Related structure data |  8d9kMC  8d58C  8d59C  8d5bC  8d9lC  8eg0C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_27264.map.gz / Format: CCP4 / Size: 11.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_27264.map.gz / Format: CCP4 / Size: 11.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Human METTL1-WDR4 proteins in complex with Lys tRNA. | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.245 Å | ||||||||||||||||||||||||||||||||||||
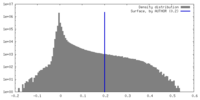
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_27264_msk_1.map emd_27264_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Human METTL1-WDR4 proteins in complex with Lys tRNA. Sharpen map.
| File | emd_27264_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Human METTL1-WDR4 proteins in complex with Lys tRNA. Sharpen map. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Human METTL1-WDR4 proteins in complex with Lys tRNA. Half map.
| File | emd_27264_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Human METTL1-WDR4 proteins in complex with Lys tRNA. Half map. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Human METTL1-WDR4 proteins in complex with Lys tRNA. Half map.
| File | emd_27264_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Human METTL1-WDR4 proteins in complex with Lys tRNA. Half map. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Human METTL1-WDR4 in complex with Lys-tRNA
| Entire | Name: Human METTL1-WDR4 in complex with Lys-tRNA |
|---|---|
| Components |
|
-Supramolecule #1: Human METTL1-WDR4 in complex with Lys-tRNA
| Supramolecule | Name: Human METTL1-WDR4 in complex with Lys-tRNA / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 112 KDa |
-Macromolecule #1: tRNA (guanine-N(7)-)-methyltransferase
| Macromolecule | Name: tRNA (guanine-N(7)-)-methyltransferase / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO / EC number: tRNA (guanine46-N7)-methyltransferase |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 31.516012 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MAAETRNVAG AEAPPPQKRY YRQRAHSNPM ADHTLRYPVK PEEMDWSELY PEFFAPLTQN QSHDDPKDKK EKRAQAQVEF ADIGCGYGG LLVELSPLFP DTLILGLEIR VKVSDYVQDR IRALRAAPAG GFQNIACLRS NAMKHLPNFF YKGQLTKMFF L FPDPHFKR ...String: MAAETRNVAG AEAPPPQKRY YRQRAHSNPM ADHTLRYPVK PEEMDWSELY PEFFAPLTQN QSHDDPKDKK EKRAQAQVEF ADIGCGYGG LLVELSPLFP DTLILGLEIR VKVSDYVQDR IRALRAAPAG GFQNIACLRS NAMKHLPNFF YKGQLTKMFF L FPDPHFKR TKHKWRIISP TLLAEYAYVL RVGGLVYTIT DVLELHDWMC THFEEHPLFE RVPLEDLSED PVVGHLGTST EE GKKVLRN GGKNFPAIFR RIQDPVLQAV TSQTSLPGH UniProtKB: tRNA (guanine-N(7)-)-methyltransferase |
-Macromolecule #2: tRNA (guanine-N(7)-)-methyltransferase non-catalytic subunit WDR4
| Macromolecule | Name: tRNA (guanine-N(7)-)-methyltransferase non-catalytic subunit WDR4 type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 45.123023 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MHHHHHHENL YFQGSGMAGS VGLALCGQTL VVRGGSRFLA TSIASSDDDS LFIYDCSAAE KKSQENKGED APLDQGSGAI LASTFSKSG SYFALTDDSK RLILFRTKPW QCLSVRTVAR RCTALTFIAS EEKVLVADKS GDVYSFSVLE PHGCGRLELG H LSMLLDVA ...String: MHHHHHHENL YFQGSGMAGS VGLALCGQTL VVRGGSRFLA TSIASSDDDS LFIYDCSAAE KKSQENKGED APLDQGSGAI LASTFSKSG SYFALTDDSK RLILFRTKPW QCLSVRTVAR RCTALTFIAS EEKVLVADKS GDVYSFSVLE PHGCGRLELG H LSMLLDVA VSPDDRFILT ADRDEKIRVS WAAAPHSIES FCLGHTEFVS RISVVPTQPG LLLSSSGDGT LRLWEYRSGR QL HCCHLAS LQELVDPQAP QKFAASRIAF WCQENCVALL CDGTPVVYIF QLDARRQQLV YRQQLAFQHQ VWDVAFEETQ GLW VLQDCQ EAPLVLYRPV GDQWQSVPES TVLKKVSGVL RGNWAMLEGS AGADASFSSL YKATFDNVTS YLKKKEERLQ QQLE KKQRR UniProtKB: tRNA (guanine-N(7)-)-methyltransferase non-catalytic subunit WDR4 |
-Macromolecule #3: RNA (65-MER)
| Macromolecule | Name: RNA (65-MER) / type: rna / ID: 3 / Number of copies: 1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 23.162705 KDa |
| Sequence | String: GCCCGGAUAG CUCAGUCGGU AGAGCAUCAG ACUUUUAAUC UGAGGGUCCA GGGUUCAAGU CCCUGUUCGG GC GENBANK: GENBANK: MN296912.1 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 3 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Mesh: 200 / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 60 sec. |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 273.15 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 62.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.2 µm / Nominal defocus min: 0.7000000000000001 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller







 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)