+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | gRAMP-TPR-CHAT (Craspase) | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | CRISPR / GRAMP / RNA BINDING PROTEIN / Craspase / RNA BINDING PROTEIN-RNA complex | |||||||||
| Function / homology | CHAT domain / CHAT domain / : / CRISPR type III-associated protein / RAMP superfamily / defense response to virus / RAMP superfamily protein / CHAT domain protein Function and homology information Function and homology information | |||||||||
| Biological species |  Candidatus Scalindua brodae (bacteria) Candidatus Scalindua brodae (bacteria) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.71 Å | |||||||||
 Authors Authors | Hu C / Nam KH / Schuler G / Ke A | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Science / Year: 2022 Journal: Science / Year: 2022Title: Craspase is a CRISPR RNA-guided, RNA-activated protease. Authors: Chunyi Hu / Sam P B van Beljouw / Ki Hyun Nam / Gabriel Schuler / Fran Ding / Yanru Cui / Alicia Rodríguez-Molina / Anna C Haagsma / Menno Valk / Martin Pabst / Stan J J Brouns / Ailong Ke /    Abstract: The CRISPR-Cas type III-E RNA-targeting effector complex gRAMP/Cas7-11 is associated with a caspase-like protein (TPR-CHAT/Csx29) to form Craspase (CRISPR-guided caspase). Here, we use cryo-electron ...The CRISPR-Cas type III-E RNA-targeting effector complex gRAMP/Cas7-11 is associated with a caspase-like protein (TPR-CHAT/Csx29) to form Craspase (CRISPR-guided caspase). Here, we use cryo-electron microscopy snapshots of Craspase to explain its target RNA cleavage and protease activation mechanisms. Target-guide pairing extending into the 5' region of the guide RNA displaces a gating loop in gRAMP, which triggers an extensive conformational relay that allosterically aligns the protease catalytic dyad and opens an amino acid side-chain-binding pocket. We further define Csx30 as the endogenous protein substrate that is site-specifically proteolyzed by RNA-activated Craspase. This protease activity is switched off by target RNA cleavage by gRAMP and is not activated by RNA targets containing a matching protospacer flanking sequence. We thus conclude that Craspase is a target RNA-activated protease with self-regulatory capacity. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_27260.map.gz emd_27260.map.gz | 22.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-27260-v30.xml emd-27260-v30.xml emd-27260.xml emd-27260.xml | 20 KB 20 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_27260.png emd_27260.png | 126.3 KB | ||
| Filedesc metadata |  emd-27260.cif.gz emd-27260.cif.gz | 7.2 KB | ||
| Others |  emd_27260_half_map_1.map.gz emd_27260_half_map_1.map.gz emd_27260_half_map_2.map.gz emd_27260_half_map_2.map.gz | 43 MB 43 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-27260 http://ftp.pdbj.org/pub/emdb/structures/EMD-27260 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27260 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27260 | HTTPS FTP |
-Validation report
| Summary document |  emd_27260_validation.pdf.gz emd_27260_validation.pdf.gz | 942.9 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_27260_full_validation.pdf.gz emd_27260_full_validation.pdf.gz | 942.5 KB | Display | |
| Data in XML |  emd_27260_validation.xml.gz emd_27260_validation.xml.gz | 11.6 KB | Display | |
| Data in CIF |  emd_27260_validation.cif.gz emd_27260_validation.cif.gz | 13.7 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-27260 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-27260 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-27260 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-27260 | HTTPS FTP |
-Related structure data
| Related structure data |  8d9fMC  8d8nC  8d97C  8d9eC  8d9gC  8d9hC  8d9iC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_27260.map.gz / Format: CCP4 / Size: 46.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_27260.map.gz / Format: CCP4 / Size: 46.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.07 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_27260_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
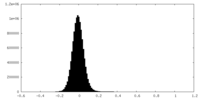
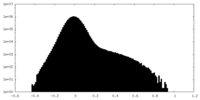
| Density Histograms |
-Half map: #1
| File | emd_27260_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
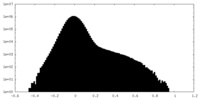
| Density Histograms |
- Sample components
Sample components
-Entire : gRAMP-TPR-CHAT Craspase
| Entire | Name: gRAMP-TPR-CHAT Craspase |
|---|---|
| Components |
|
-Supramolecule #1: gRAMP-TPR-CHAT Craspase
| Supramolecule | Name: gRAMP-TPR-CHAT Craspase / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 |
|---|---|
| Source (natural) | Organism:  Candidatus Scalindua brodae (bacteria) Candidatus Scalindua brodae (bacteria) |
-Macromolecule #1: CHAT domain protein
| Macromolecule | Name: CHAT domain protein / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Candidatus Scalindua brodae (bacteria) Candidatus Scalindua brodae (bacteria) |
| Molecular weight | Theoretical: 77.26343 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: TREDIDRKEA ERLLDEAFNP RTKPVDRKKI INSALKILIG LYKEKKDDLT SASFISIARA YYLVSITILP KGTTIPEKKK EALRKGIEF IDRAINKFNG SILDSQRAFR IKSVLSIEFN RIDREKCDNI KLKNLLNEAV DKGCTDFDTY EWDIQIAIRL C ELGVDMEG ...String: TREDIDRKEA ERLLDEAFNP RTKPVDRKKI INSALKILIG LYKEKKDDLT SASFISIARA YYLVSITILP KGTTIPEKKK EALRKGIEF IDRAINKFNG SILDSQRAFR IKSVLSIEFN RIDREKCDNI KLKNLLNEAV DKGCTDFDTY EWDIQIAIRL C ELGVDMEG HFDNLIKSNK ANDLQKAKAY YFIKKDDHKA KEHMDKCTAS LKYTPCSHRL WDETVGFIER LKGDSSTLWR DF AIKTYRS CRVQEKETGT LRLRWYWSRH RVLYDMAFLA VKEQADVNVK QAKIKKLAEI SDSLKSRFSL RLSDMEKMPK SDD ESNHEF KKFLDKCVTA YQDGYVILLE LTQVPEGWVV VHFYLNKLEG MGNAIVFDKC ANSWQYKEFQ YKELFEVFLT WQAN YNLYK ENAAEHLVTL CKKIGETMPF LFCDNFIPNG KDVLFVPHDF LHRLPLHGSI ENKTNGKLFL ENHSCCYLPA WSFAS EKEA STSDEYVLLK NFDQGHFETL QNNQIWGTQS VKDGASSDDL ENIRNNPRLL TILCHGEANM SNPFRSMLKL ANGGIT YLE ILNSVKGLKG SQVILGACET DLVPPLSDVM DEHYSVATAL LLIGAAGVVG TMWKVRSNKT KSLIEWKLEN IEYKLNE WQ KETGGAAYKD HPPTFYRSIA FRSIGFPL UniProtKB: CHAT domain protein |
-Macromolecule #2: RAMP superfamily protein
| Macromolecule | Name: RAMP superfamily protein / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Candidatus Scalindua brodae (bacteria) Candidatus Scalindua brodae (bacteria) |
| Molecular weight | Theoretical: 142.320609 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MNITVELTFF EPYRLVEWFD WDARKKSHSA MRGQAFAQWT WKGKGRTAGK SFITGTLVRS AVIKAVEELL SLNNGKWEGV PCCNGSFQT DESKGKKPSF LRKRHTLQWQ ANNKNICDKE EACPFCILLG RFDNAGKVHE RNKDYDIHFS NFDLDHDLRL V DIASGRIL ...String: MNITVELTFF EPYRLVEWFD WDARKKSHSA MRGQAFAQWT WKGKGRTAGK SFITGTLVRS AVIKAVEELL SLNNGKWEGV PCCNGSFQT DESKGKKPSF LRKRHTLQWQ ANNKNICDKE EACPFCILLG RFDNAGKVHE RNKDYDIHFS NFDLDHDLRL V DIASGRIL NRVDFDTGKA KDYFRTWEAD YETYGTYTGR ITLRNEHAKK LLLASLGFVD KLCGALCRIE VIKSEDHNDE LR KQAEVIV EAFKQNDKLE KIRILADAIR TLRLHGEGVI EKDELPDGKE ERDKGHHLWD IKVQGTALRT KLKELWQSNK DIG WRKFTE MLGSNLYLIY KKETGGVSTR FRILGDTEYY SKAHDSEGSD LFIPVTPPEG IETKEWIIVG RLKAATPFYF GVQQ PSDSI PGKEKKSEDS LVINEHTSFN ILLDKENRYR IPRSALRGAL RRDLRTAFGS GCNVSLGGQI LCNCKVCIEM RRITL KDSV SDFSEPPEIR YRIAKNPGTA TVEDGSLFDI EVGPEGLTFP FVLRYRGHKF PEQLSSVIRY WEENDGKNGM AWLGGL DST GKGRFALKDI KIFEWDLNQK INEYIKERGM RGKEKELLEM GESSLPDGLI PYKFFEEREC LFPYKENLKP QWSEVQY TI EVGSPLLTAD TISALTEPGN RDAIAYKKRV YNDGNNAIEP EPRFAVKSET HRGIFRTAVG RRTGDLGKED HEDCTCDM C IIFGNEHESS KIRFEDLELI NGNEFEKLEK HIDHVAIDRF TGGALDKAKF DTYPLAGSPK KPLKLKGRFW IKKGFSGDH KLLITTALSD IRDGLYPLGS KGGVGYGWVA GISIDDNVPD DFKEMINKTY VHPGHQSPKQ DHKNKNIYYP HYFLDSGSKV YREKDIITH EEFTEELLSG KINCKLETLT PLIIPDTSDE NGLKLQGNKP GHKNYKFFNI NGELMIPGSE LRGMLRTHFE A LTKSCFAI FGEGGKLDKA LHPCTGLSDG LCPGCHLFGT TDYKGRVKFG FAKYENGPEW LITRGNNPER SLTLGVLESP RP AFSIPDD ESEIPGRKFY LHHNGWRIIR QKQLEIRETV QPERNVTTEV MDKGNVFSFD VRFENLREWE LGLLLQSLDP GKN IAHKLG KGKPYGFGSV KIKIDSLHTF KIIKRVPQSD IREYINKGYQ KLIEWSLPQW HVIPHIDKLY KLLWVPFLND SKLE PDVRY PVLNYTYKKL GDKDNLPYKT RVKGLTTPWS PWNPFQV UniProtKB: RAMP superfamily protein, RAMP superfamily protein |
-Macromolecule #3: RNA (33-MER)
| Macromolecule | Name: RNA (33-MER) / type: rna / ID: 3 / Number of copies: 1 |
|---|---|
| Source (natural) | Organism:  Candidatus Scalindua brodae (bacteria) Candidatus Scalindua brodae (bacteria) |
| Molecular weight | Theoretical: 10.404171 KDa |
| Sequence | String: GACUUAAUGU CACGGUACCC AAUUUUCUGC CCC |
-Macromolecule #4: ZINC ION
| Macromolecule | Name: ZINC ION / type: ligand / ID: 4 / Number of copies: 4 / Formula: ZN |
|---|---|
| Molecular weight | Theoretical: 65.409 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 59.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)