+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Plasmodium falciparum merozoite with a fused rhoptry pair | |||||||||
 Map data Map data | Plasmodium falciparum merozoite with a fused rhoptry pair | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Rhoptry / Parasite / CELL INVASION | |||||||||
| Biological species |  | |||||||||
| Method | electron tomography / cryo EM | |||||||||
 Authors Authors | Martinez M / Chang Y-W | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Microbiol / Year: 2022 Journal: Nat Microbiol / Year: 2022Title: Rhoptry secretion system structure and priming in Plasmodium falciparum revealed using in situ cryo-electron tomography. Authors: Matthew Martinez / William David Chen / Marta Mendonça Cova / Petra Molnár / Shrawan Kumar Mageswaran / Amandine Guérin / Audrey R Odom John / Maryse Lebrun / Yi-Wei Chang /   Abstract: Apicomplexan parasites secrete contents of the rhoptries, club-shaped organelles in the apical region, into host cells to permit their invasion and establishment of infection. The rhoptry secretory ...Apicomplexan parasites secrete contents of the rhoptries, club-shaped organelles in the apical region, into host cells to permit their invasion and establishment of infection. The rhoptry secretory apparatus (RSA), which is critical for rhoptry secretion, was recently discovered in Toxoplasma and Cryptosporidium. It is unknown whether a similar molecular machinery exists in the malaria parasite Plasmodium. In this study, we use in situ cryo-electron tomography to investigate the rhoptry secretion system in P. falciparum merozoites. We identify the presence of an RSA at the cell apex and a morphologically distinct apical vesicle docking the tips of the two rhoptries to the RSA. We also discover two additional rhoptry organizations that lack the apical vesicle. Using subtomogram averaging, we reveal different conformations of the RSA structure corresponding to different rhoptry organizations. Our results highlight previously unknown steps in the process of rhoptry secretion and indicate a regulatory role for the conserved apical vesicle in host invasion by apicomplexan parasites. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_26747.map.gz emd_26747.map.gz | 1.5 GB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-26747-v30.xml emd-26747-v30.xml emd-26747.xml emd-26747.xml | 10.4 KB 10.4 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_26747.png emd_26747.png | 196.2 KB | ||
| Filedesc metadata |  emd-26747.cif.gz emd-26747.cif.gz | 4.2 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-26747 http://ftp.pdbj.org/pub/emdb/structures/EMD-26747 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26747 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26747 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_26747.map.gz / Format: CCP4 / Size: 2.2 GB / Type: IMAGE STORED AS SIGNED INTEGER (2 BYTES) Download / File: emd_26747.map.gz / Format: CCP4 / Size: 2.2 GB / Type: IMAGE STORED AS SIGNED INTEGER (2 BYTES) | ||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Plasmodium falciparum merozoite with a fused rhoptry pair | ||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. generated in cubic-lattice coordinate | ||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 10.6 Å | ||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
- Sample components
Sample components
-Entire : Plasmodium falciparum merozoite apical complex
| Entire | Name: Plasmodium falciparum merozoite apical complex |
|---|---|
| Components |
|
-Supramolecule #1: Plasmodium falciparum merozoite apical complex
| Supramolecule | Name: Plasmodium falciparum merozoite apical complex / type: cell / ID: 1 / Parent: 0 Details: Two separate rhoptries lacking an associated apical vesicle |
|---|---|
| Source (natural) | Organism:  |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | electron tomography |
| Aggregation state | cell |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 Details: RPMI-1640 supplemented with 27 mM NaHCO3, 11 mM glucose, 5 mM HEPES, 0.01 mM thymidine, 1 mM sodium pyruvate, 0.37 mM hypoxanthine, 10 ug/mL gentamicin |
|---|---|
| Grid | Model: Quantifoil R2/2 / Material: COPPER / Mesh: 200 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE-PROPANE / Chamber humidity: 95 % / Chamber temperature: 297 K / Instrument: LEICA EM GP / Details: Front blot for 4-6 seconds before plunging. |
| Details | Mechanically isolated merozoites from synchronized schizonts |
| Sectioning | Other: NO SECTIONING |
| Fiducial marker | Manufacturer: Ted Pella, Inc / Diameter: 10 nm |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Phase plate: VOLTA PHASE PLATE / Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average exposure time: 0.5 sec. / Average electron dose: 140.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 5.0 µm / Nominal defocus min: 1.5 µm / Nominal magnification: 33000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Details | Raw frames were gain normalized in serialEM and motion corrected using the alignframes command in IMOD |
|---|---|
| Final reconstruction | Algorithm: BACK PROJECTION / Software - Name:  IMOD / Number images used: 61 IMOD / Number images used: 61 |
 Movie
Movie Controller
Controller



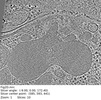



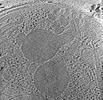
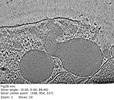
 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)
















