[English] 日本語
 Yorodumi
Yorodumi- EMDB-26704: Cryo-EM Structure of the Neutralizing Antibody MPV467 in Complex ... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM Structure of the Neutralizing Antibody MPV467 in Complex with Prefusion Human Metapneumovirus F Glycoprotein | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | neutralizing antibody / fusion protein / metapneumovirus / VIRAL PROTEIN-IMMUNE SYSTEM complex | |||||||||
| Function / homology | Precursor fusion glycoprotein F0, Paramyxoviridae / Fusion glycoprotein F0 / fusion of virus membrane with host plasma membrane / viral envelope / symbiont entry into host cell / host cell plasma membrane / virion membrane / membrane / Fusion glycoprotein F0 Function and homology information Function and homology information | |||||||||
| Biological species |  Human metapneumovirus / Human metapneumovirus /  Homo sapiens (human) Homo sapiens (human) | |||||||||
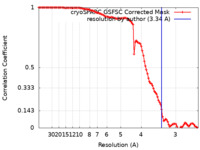
| Method | single particle reconstruction / cryo EM / Resolution: 3.34 Å | |||||||||
 Authors Authors | Rush SA / McLellan JS | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2022 Journal: Proc Natl Acad Sci U S A / Year: 2022Title: Structural basis for ultrapotent antibody-mediated neutralization of human metapneumovirus. Authors: Avik Banerjee / Jiachen Huang / Scott A Rush / Jackelyn Murray / Aaron D Gingerich / Fredejah Royer / Ching-Lin Hsieh / Ralph A Tripp / Jason S McLellan / Jarrod J Mousa /  Abstract: Human metapneumovirus (hMPV) is a leading cause of morbidity and hospitalization among children worldwide, however, no vaccines or therapeutics are currently available for hMPV disease prevention and ...Human metapneumovirus (hMPV) is a leading cause of morbidity and hospitalization among children worldwide, however, no vaccines or therapeutics are currently available for hMPV disease prevention and treatment. The hMPV fusion (F) protein is the sole target of neutralizing antibodies. To map the immunodominant epitopes on the hMPV F protein, we isolated a panel of human monoclonal antibodies (mAbs), and the mAbs were assessed for binding avidity, neutralization potency, and epitope specificity. We found the majority of the mAbs target diverse epitopes on the hMPV F protein, and we discovered multiple mAb binding approaches for antigenic site III. The most potent mAb, MPV467, which had picomolar potency, was examined in prophylactic and therapeutic mouse challenge studies, and MPV467 limited virus replication in mouse lungs when administered 24 h before or 72 h after viral infection. We determined the structure of MPV467 in complex with the hMPV F protein using cryo-electron microscopy to a resolution of 3.3 Å, which revealed a complex novel prefusion-specific epitope overlapping antigenic sites II and V on a single protomer. Overall, our data reveal insights into the immunodominant antigenic epitopes on the hMPV F protein, identify a mAb therapy for hMPV F disease prevention and treatment, and provide the discovery of a prefusion-specific epitope on the hMPV F protein. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_26704.map.gz emd_26704.map.gz | 63 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-26704-v30.xml emd-26704-v30.xml emd-26704.xml emd-26704.xml | 21.2 KB 21.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_26704_fsc.xml emd_26704_fsc.xml | 11.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_26704.png emd_26704.png | 81.3 KB | ||
| Masks |  emd_26704_msk_1.map emd_26704_msk_1.map | 125 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-26704.cif.gz emd-26704.cif.gz | 6.8 KB | ||
| Others |  emd_26704_additional_1.map.gz emd_26704_additional_1.map.gz emd_26704_half_map_1.map.gz emd_26704_half_map_1.map.gz emd_26704_half_map_2.map.gz emd_26704_half_map_2.map.gz | 107.8 MB 116.1 MB 116.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-26704 http://ftp.pdbj.org/pub/emdb/structures/EMD-26704 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26704 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26704 | HTTPS FTP |
-Validation report
| Summary document |  emd_26704_validation.pdf.gz emd_26704_validation.pdf.gz | 821.1 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_26704_full_validation.pdf.gz emd_26704_full_validation.pdf.gz | 820.7 KB | Display | |
| Data in XML |  emd_26704_validation.xml.gz emd_26704_validation.xml.gz | 19.2 KB | Display | |
| Data in CIF |  emd_26704_validation.cif.gz emd_26704_validation.cif.gz | 24.9 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26704 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26704 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26704 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26704 | HTTPS FTP |
-Related structure data
| Related structure data |  7ur4MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_26704.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_26704.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.269 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_26704_msk_1.map emd_26704_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: #1
| File | emd_26704_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_26704_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_26704_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Trimeric prefusion hMPV F glycoprotein bound by three molecules o...
| Entire | Name: Trimeric prefusion hMPV F glycoprotein bound by three molecules of MPV467 Fab |
|---|---|
| Components |
|
-Supramolecule #1: Trimeric prefusion hMPV F glycoprotein bound by three molecules o...
| Supramolecule | Name: Trimeric prefusion hMPV F glycoprotein bound by three molecules of MPV467 Fab type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 |
|---|---|
| Source (natural) | Organism:  Human metapneumovirus / Strain: NL/1/00 Human metapneumovirus / Strain: NL/1/00 |
-Macromolecule #1: Fusion glycoprotein F0
| Macromolecule | Name: Fusion glycoprotein F0 / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Human metapneumovirus Human metapneumovirus |
| Molecular weight | Theoretical: 60.48993 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MSWKVVIIFS LLITPQHGLK ESYLEESCST ITEGYLSVLR TGWYTNVFTL EVGDVENLTC ADGPSLIKTE LDLTKSALRE LRTCSADQL AREEQIENPR RRRFVLGAIA CGVATAAAVT AGVAIAKCIR LESEVTAIKN CLKKTNECVS TLGCGVRVLA T AVRELKDF ...String: MSWKVVIIFS LLITPQHGLK ESYLEESCST ITEGYLSVLR TGWYTNVFTL EVGDVENLTC ADGPSLIKTE LDLTKSALRE LRTCSADQL AREEQIENPR RRRFVLGAIA CGVATAAAVT AGVAIAKCIR LESEVTAIKN CLKKTNECVS TLGCGVRVLA T AVRELKDF VSKNLTRAIN KNKCDIPDLK MAVSFSQFNR RFLNVVRQFS DNAGITPAIS KDLMTDAELA RAISNMPTSA GQ IKLMLEN RCMVRRKGFG ILIGVYGSSV IYMVQLPIFG VIDTPCWIVK AAPSCSEKKG NYACLLREDQ GWYCQNAGST VYY PCEKDC ETRGDHVFCD TAAGINVAEQ SKECNINIST TNYPCKVSCG RHPISMVALS PLGALVACYK GVSCSIGSNR VGII KQLNK GCSYITNQDA DTVTIDNTVY QLSKVEGEQH VIKGRPVSSS FDPVKFPQDQ FNVALDQCFE SIENSQALVD QSNRI LSSA EKGNTGGGGS GYIPEAPRDG QAYVRKDGEW VLLSTFLGRS LEVLFQGPGH HHHHHHHSAW SHPQFEK UniProtKB: Fusion glycoprotein F0 |
-Macromolecule #2: MPV467 Fab Heavy chain
| Macromolecule | Name: MPV467 Fab Heavy chain / type: protein_or_peptide / ID: 2 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 24.023779 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: LVQSGGGVVR PGTSLRVSCA AFDFNFRDYG MHWVRQAPGK GLEWVAGIWY DGSNKDYADS VKGRFTISRD NSQNTLYLQM NSLRVEDTA VYYCARDPRT HREGALSHFD SWGQGTLVTV SSASTKGPSV FPLAPSSKST SGGTAALGCL VKDYFPEPVT V SWNSGALT ...String: LVQSGGGVVR PGTSLRVSCA AFDFNFRDYG MHWVRQAPGK GLEWVAGIWY DGSNKDYADS VKGRFTISRD NSQNTLYLQM NSLRVEDTA VYYCARDPRT HREGALSHFD SWGQGTLVTV SSASTKGPSV FPLAPSSKST SGGTAALGCL VKDYFPEPVT V SWNSGALT SGVHTFPAVL QSSGLYSLSS VVTVPSSSLG TQTYICNVNH KPSNTKVDKR VEPKS |
-Macromolecule #3: MPV467 Fab Light chain
| Macromolecule | Name: MPV467 Fab Light chain / type: protein_or_peptide / ID: 3 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 22.621924 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: ELTQDPAVSV ALGQTVRITC QGDSLRNYFA GWYQQKPGQA PLLVLYGENI RPSGIPDRFS GSSSGNTVSL TITGAQAEDE ADYYCNSRD NSGNHWVFGG GTRLTVLGQP KAAPSVTLFP PSSEELQANK ATLVCLISDF YPGAVTVAWK ADSSPVKAGV E TTTPSKQS ...String: ELTQDPAVSV ALGQTVRITC QGDSLRNYFA GWYQQKPGQA PLLVLYGENI RPSGIPDRFS GSSSGNTVSL TITGAQAEDE ADYYCNSRD NSGNHWVFGG GTRLTVLGQP KAAPSVTLFP PSSEELQANK ATLVCLISDF YPGAVTVAWK ADSSPVKAGV E TTTPSKQS NNKYAASSYL SLTPEQWKSH RSYSCQVTHE GSTVEKTVAP TEC |
-Macromolecule #5: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 5 / Number of copies: 6 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 Component:
| |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Grid | Model: C-flat-1.2/1.3 / Material: GOLD / Mesh: 300 / Support film - Material: GOLD / Support film - topology: HOLEY / Pretreatment - Type: PLASMA CLEANING / Pretreatment - Time: 180 sec. / Pretreatment - Atmosphere: OTHER | |||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 283.15 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | TFS GLACIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 150000 |
| Sample stage | Cooling holder cryogen: NITROGEN |
 Movie
Movie Controller
Controller





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)