[English] 日本語
 Yorodumi
Yorodumi- EMDB-26438: Transcription antitermination complex: "pre-engaged" Qlambda-load... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Transcription antitermination complex: "pre-engaged" Qlambda-loading complex | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | RNA polymerase / DNA Binding / transcription / Q-dependent antitermination / Q antitermination factor / GENE REGULATION / TRANSFERASE-DNA-RNA complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationsubmerged biofilm formation / cellular response to cell envelope stress / regulation of DNA-templated transcription initiation / sigma factor activity / bacterial-type flagellum assembly / cytosolic DNA-directed RNA polymerase complex / bacterial-type flagellum-dependent cell motility / nitrate assimilation / DNA-directed RNA polymerase complex / regulation of DNA-templated transcription elongation ...submerged biofilm formation / cellular response to cell envelope stress / regulation of DNA-templated transcription initiation / sigma factor activity / bacterial-type flagellum assembly / cytosolic DNA-directed RNA polymerase complex / bacterial-type flagellum-dependent cell motility / nitrate assimilation / DNA-directed RNA polymerase complex / regulation of DNA-templated transcription elongation / transcription elongation factor complex / transcription antitermination / cell motility / DNA-templated transcription initiation / DNA-templated transcription termination / ribonucleoside binding / DNA-directed RNA polymerase / DNA-directed RNA polymerase activity / response to heat / intracellular iron ion homeostasis / protein dimerization activity / response to antibiotic / DNA-templated transcription / magnesium ion binding / DNA binding / zinc ion binding / metal ion binding / membrane / cytosol / cytoplasm Similarity search - Function | |||||||||
| Biological species |   Escherichia phage Lambda (virus) Escherichia phage Lambda (virus) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.13 Å | |||||||||
 Authors Authors | Yin Z / Ebright RH | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2022 Journal: Proc Natl Acad Sci U S A / Year: 2022Title: In transcription antitermination by Qλ, NusA induces refolding of Qλ to form a nozzle that extends the RNA polymerase RNA-exit channel. Authors: Zhou Yin / Jeremy G Bird / Jason T Kaelber / Bryce E Nickels / Richard H Ebright /  Abstract: Lambdoid bacteriophage Q proteins are transcription antipausing and antitermination factors that enable RNA polymerase (RNAP) to read through pause and termination sites. Q proteins load onto RNAP ...Lambdoid bacteriophage Q proteins are transcription antipausing and antitermination factors that enable RNA polymerase (RNAP) to read through pause and termination sites. Q proteins load onto RNAP engaged in promoter-proximal pausing at a Q binding element (QBE) and adjacent sigma-dependent pause element to yield a Q-loading complex, and they translocate with RNAP as a pausing-deficient, termination-deficient Q-loaded complex. In previous work, we showed that the Q protein of bacteriophage 21 (Q21) functions by forming a nozzle that narrows and extends the RNAP RNA-exit channel, preventing formation of pause and termination RNA hairpins. Here, we report atomic structures of four states on the pathway of antitermination by the Q protein of bacteriophage λ (Qλ), a Q protein that shows no sequence similarity to Q21 and that, unlike Q21, requires the transcription elongation factor NusA for efficient antipausing and antitermination. We report structures of Qλ, the Qλ-QBE complex, the NusA-free pre-engaged Qλ-loading complex, and the NusA-containing engaged Qλ-loading complex. The results show that Qλ, like Q21, forms a nozzle that narrows and extends the RNAP RNA-exit channel, preventing formation of RNA hairpins. However, the results show that Qλ has no three-dimensional structural similarity to Q21, employs a different mechanism of QBE recognition than Q21, and employs a more complex process for loading onto RNAP than Q21, involving recruitment of Qλ to form a pre-engaged loading complex, followed by NusA-facilitated refolding of Qλ to form an engaged loading complex. The results establish that Qλ and Q21 are not structural homologs and are solely functional analogs. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_26438.map.gz emd_26438.map.gz | 39.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-26438-v30.xml emd-26438-v30.xml emd-26438.xml emd-26438.xml | 30.4 KB 30.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_26438_fsc.xml emd_26438_fsc.xml | 9.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_26438.png emd_26438.png | 147.8 KB | ||
| Filedesc metadata |  emd-26438.cif.gz emd-26438.cif.gz | 9.5 KB | ||
| Others |  emd_26438_half_map_1.map.gz emd_26438_half_map_1.map.gz emd_26438_half_map_2.map.gz emd_26438_half_map_2.map.gz | 49.7 MB 49.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-26438 http://ftp.pdbj.org/pub/emdb/structures/EMD-26438 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26438 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26438 | HTTPS FTP |
-Related structure data
| Related structure data |  7ubmMC  7ubjC  7ubkC 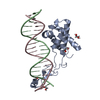 7ublC  7ubnC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_26438.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_26438.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.066 Å | ||||||||||||||||||||||||||||||||||||
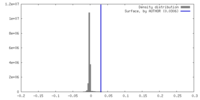
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_26438_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
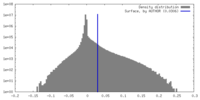
| Density Histograms |
-Half map: #2
| File | emd_26438_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
+Entire : Transcription antitermination complex: "pre-engaged" Qlambda-load...
+Supramolecule #1: Transcription antitermination complex: "pre-engaged" Qlambda-load...
+Macromolecule #1: DNA (53-MER)
+Macromolecule #2: DNA (52-MER)
+Macromolecule #3: DNA-directed RNA polymerase subunit alpha
+Macromolecule #4: DNA-directed RNA polymerase subunit beta
+Macromolecule #5: DNA-directed RNA polymerase subunit beta'
+Macromolecule #6: DNA-directed RNA polymerase subunit omega
+Macromolecule #7: RNA polymerase sigma factor RpoD
+Macromolecule #8: Antitermination protein Q
+Macromolecule #9: RNA (5'-R(*UP*GP*GP*GP*AP*GP*AP*GP*GP*UP*A)-3')
+Macromolecule #10: ZINC ION
+Macromolecule #11: MAGNESIUM ION
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 9 mg/mL | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.9 Component:
| |||||||||||||||
| Grid | Model: UltrAuFoil R1.2/1.3 / Material: GOLD / Mesh: 300 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 300 sec. | |||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 292 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Number grids imaged: 1 / Number real images: 1643 / Average exposure time: 5.0 sec. / Average electron dose: 1.41 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.5 µm / Nominal defocus min: 0.8 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model |
| ||||||
|---|---|---|---|---|---|---|---|
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT / Overall B value: 100 / Target criteria: Correlation coefficient | ||||||
| Output model |  PDB-7ubm: |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)