+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of human ankyrin complex (B2P1A1)2 | |||||||||
 Map data Map data | Cryo-EM structure of Ankyrin complex (B2P1A1)2 | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Red blood cell / Ankyrin complex / membrane protein / band 3 / protein 4.2 | |||||||||
| Function / homology |  Function and homology information Function and homology informationspectrin-associated cytoskeleton / hemoglobin metabolic process / positive regulation of organelle organization / maintenance of epithelial cell apical/basal polarity / protein-glutamine gamma-glutamyltransferase activity / pH elevation / Defective SLC4A1 causes hereditary spherocytosis type 4 (HSP4), distal renal tubular acidosis (dRTA) and dRTA with hemolytic anemia (dRTA-HA) / negative regulation of urine volume / NrCAM interactions / Neurofascin interactions ...spectrin-associated cytoskeleton / hemoglobin metabolic process / positive regulation of organelle organization / maintenance of epithelial cell apical/basal polarity / protein-glutamine gamma-glutamyltransferase activity / pH elevation / Defective SLC4A1 causes hereditary spherocytosis type 4 (HSP4), distal renal tubular acidosis (dRTA) and dRTA with hemolytic anemia (dRTA-HA) / negative regulation of urine volume / NrCAM interactions / Neurofascin interactions / Bicarbonate transporters / intracellular monoatomic ion homeostasis / ankyrin-1 complex / CHL1 interactions / monoatomic anion transmembrane transporter activity / plasma membrane phospholipid scrambling / cytoskeletal anchor activity / chloride:bicarbonate antiporter activity / solute:inorganic anion antiporter activity / bicarbonate transport / bicarbonate transmembrane transporter activity / M band / monoatomic anion transport / chloride transport / chloride transmembrane transporter activity / Interaction between L1 and Ankyrins / ankyrin binding / hemoglobin binding / spectrin binding / erythrocyte maturation / erythrocyte development / negative regulation of glycolytic process through fructose-6-phosphate / cortical cytoskeleton / exocytosis / axolemma / endoplasmic reticulum to Golgi vesicle-mediated transport / spleen development / COPI-mediated anterograde transport / protein-membrane adaptor activity / cytoskeleton organization / chloride transmembrane transport / sarcoplasmic reticulum / regulation of intracellular pH / protein localization to plasma membrane / Erythrocytes take up oxygen and release carbon dioxide / Erythrocytes take up carbon dioxide and release oxygen / sarcolemma / structural constituent of cytoskeleton / multicellular organismal-level iron ion homeostasis / transmembrane transport / cytoplasmic side of plasma membrane / Z disc / cell morphogenesis / blood coagulation / regulation of cell shape / ATPase binding / protein phosphatase binding / blood microparticle / basolateral plasma membrane / postsynaptic membrane / transmembrane transporter binding / cytoskeleton / neuron projection / structural molecule activity / enzyme binding / signal transduction / protein homodimerization activity / extracellular exosome / ATP binding / membrane / plasma membrane / cytosol Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 8.5 Å | |||||||||
 Authors Authors | Xia X / Liu SH / Zhou ZH | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2022 Journal: Nat Struct Mol Biol / Year: 2022Title: Structure, dynamics and assembly of the ankyrin complex on human red blood cell membrane. Authors: Xian Xia / Shiheng Liu / Z Hong Zhou /  Abstract: The cytoskeleton of a red blood cell (RBC) is anchored to the cell membrane by the ankyrin complex. This complex is assembled during RBC genesis and comprises primarily band 3, protein 4.2 and ...The cytoskeleton of a red blood cell (RBC) is anchored to the cell membrane by the ankyrin complex. This complex is assembled during RBC genesis and comprises primarily band 3, protein 4.2 and ankyrin, whose mutations contribute to numerous human inherited diseases. High-resolution structures of the ankyrin complex have been long sought-after to understand its assembly and disease-causing mutations. Here, we analyzed native complexes on the human RBC membrane by stepwise fractionation. Cryo-electron microscopy structures of nine band-3-associated complexes reveal that protein 4.2 stabilizes the cytoplasmic domain of band 3 dimer. In turn, the superhelix-shaped ankyrin binds to this protein 4.2 via ankyrin repeats (ARs) 6-13 and to another band 3 dimer via ARs 17-20, bridging two band 3 dimers in the ankyrin complex. Integration of these structures with both prior data and our biochemical data supports a model of ankyrin complex assembly during erythropoiesis and identifies interactions essential for the mechanical stability of RBC. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_26154.map.gz emd_26154.map.gz | 41.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-26154-v30.xml emd-26154-v30.xml emd-26154.xml emd-26154.xml | 18.5 KB 18.5 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_26154.png emd_26154.png | 46 KB | ||
| Filedesc metadata |  emd-26154.cif.gz emd-26154.cif.gz | 6.9 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-26154 http://ftp.pdbj.org/pub/emdb/structures/EMD-26154 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26154 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26154 | HTTPS FTP |
-Validation report
| Summary document |  emd_26154_validation.pdf.gz emd_26154_validation.pdf.gz | 406.8 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_26154_full_validation.pdf.gz emd_26154_full_validation.pdf.gz | 406.3 KB | Display | |
| Data in XML |  emd_26154_validation.xml.gz emd_26154_validation.xml.gz | 6.6 KB | Display | |
| Data in CIF |  emd_26154_validation.cif.gz emd_26154_validation.cif.gz | 7.5 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26154 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26154 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26154 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26154 | HTTPS FTP |
-Related structure data
| Related structure data |  7tvzC  7tw0C  7tw1C  7tw2C  7tw3C  7tw5C  7tw6C C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_26154.map.gz / Format: CCP4 / Size: 83.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_26154.map.gz / Format: CCP4 / Size: 83.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryo-EM structure of Ankyrin complex (B2P1A1)2 | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.2 Å | ||||||||||||||||||||||||||||||||||||
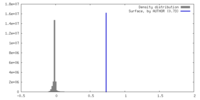
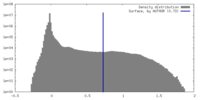
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
- Sample components
Sample components
-Entire : High-salt fraction 2 from human red blood cell membrane
| Entire | Name: High-salt fraction 2 from human red blood cell membrane |
|---|---|
| Components |
|
-Supramolecule #1: High-salt fraction 2 from human red blood cell membrane
| Supramolecule | Name: High-salt fraction 2 from human red blood cell membrane type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 1 MDa |
-Macromolecule #1: band 3
| Macromolecule | Name: band 3 / type: protein_or_peptide / ID: 1 / Details: four band 3 in the complex / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MEELQDDYED MMEENLEQEE YEDPDIPESQ MEEPAAHDTE ATATDYHTTS HPGTHKVYVE LQELVMDEKN QELRWMEAAR WVQLEENLGE NGAWGRPHLS HLTFWSLLEL RRVFTKGTVL LDLQETSLAG VANQLLDRFI FEDQIRPQDR EELLRALLLK HSHAGELEAL ...String: MEELQDDYED MMEENLEQEE YEDPDIPESQ MEEPAAHDTE ATATDYHTTS HPGTHKVYVE LQELVMDEKN QELRWMEAAR WVQLEENLGE NGAWGRPHLS HLTFWSLLEL RRVFTKGTVL LDLQETSLAG VANQLLDRFI FEDQIRPQDR EELLRALLLK HSHAGELEAL GGVKPAVLTR SGDPSQPLLP QHSSLETQLF CEQGDGGTEG HSPSGILEKI PPDSEATLVL VGRADFLEQP VLGFVRLQEA AELEAVELPV PIRFLFVLLG PEAPHIDYTQ LGRAAATLMS ERVFRIDAYM AQSRGELLHS LEGFLDCSLV LPPTDAPSEQ ALLSLVPVQR ELLRRRYQSS PAKPDSSFYK GLDLNGGPDD PLQQTGQLFG GLVRDIRRRY PYYLSDITDA FSPQVLAAVI FIYFAALSPA ITFGGLLGEK TRNQMGVSEL LISTAVQGIL FALLGAQPLL VVGFSGPLLV FEEAFFSFCE TNGLEYIVGR VWIGFWLILL VVLVVAFEGS FLVRFISRYT QEIFSFLISL IFIYETFSKL IKIFQDHPLQ KTYNYNVLMV PKPQGPLPNT ALLSLVLMAG TFFFAMMLRK FKNSSYFPGK LRRVIGDFGV PISILIMVLV DFFIQDTYTQ KLSVPDGFKV SNSSARGWVI HPLGLRSEFP IWMMFASALP ALLVFILIFL ESQITTLIVS KPERKMVKGS GFHLDLLLVV GMGGVAALFG MPWLSATTVR SVTHANALTV MGKASTPGAA AQIQEVKEQR ISGLLVAVLV GLSILMEPIL SRIPLAVLFG IFLYMGVTSL SGIQLFDRIL LLFKPPKYHP DVPYVKRVKT WRMHLFTGIQ IICLAVLWVV KSTPASLALP FVLILTVPLR RVLLPLIFRN VELQCLDADD AKATFDEEEG RDEYDEVAMP V UniProtKB: Band 3 anion transport protein |
-Macromolecule #2: protein 4.2
| Macromolecule | Name: protein 4.2 / type: protein_or_peptide / ID: 2 / Details: two protein 4.2 in the complex / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MGQALGIKSC DFQAARNNEE HHTKALSSRR LFVRRGQPFT IILYFRAPVR AFLPALKKVA LTAQTGEQPS KINRTQATFP ISSLGDRKWW SAVVEERDAQ SWTISVTTPA DAVIGHYSLL LQVSGRKQLL LGQFTLLFNP WNREDAVFLK NEAQRMEYLL NQNGLIYLGT ...String: MGQALGIKSC DFQAARNNEE HHTKALSSRR LFVRRGQPFT IILYFRAPVR AFLPALKKVA LTAQTGEQPS KINRTQATFP ISSLGDRKWW SAVVEERDAQ SWTISVTTPA DAVIGHYSLL LQVSGRKQLL LGQFTLLFNP WNREDAVFLK NEAQRMEYLL NQNGLIYLGT ADCIQAESWD FGQFEGDVID LSLRLLSKDK QVEKWSQPVH VARVLGALLH FLKEQRVLPT PQTQATQEGA LLNKRRGSVP ILRQWLTGRG RPVYDGQAWV LAAVACTVLR CLGIPARVVT TFASAQGTGG RLLIDEYYNE EGLQNGEGQR GRIWIFQTST ECWMTRPALP QGYDGWQILH PSAPNGGGVL GSCDLVPVRA VKEGTLGLTP AVSDLFAAIN ASCVVWKCCE DGTLELTDSN TKYVGNNIST KGVGSDRCED ITQNYKYPEG SLQEKEVLER VEKEKMEREK DNGIRPPSLE TASPLYLLLK APSSLPLRGD AQISVTLVNH SEQEKAVQLA IGVQAVHYNG VLAAKLWRKK LHLTLSANLE KIITIGLFFS NFERNPPENT FLRLTAMATH SESNLSCFAQ EDIAICRPHL AIKMPEKAEQ YQPLTASVSL QNSLDAPMED CVISILGRGL IHRERSYRFR SVWPENTMCA KFQFTPTHVG LQRLTVEVDC NMFQNLTNYK SVTVVAPELS A UniProtKB: Protein 4.2 |
-Macromolecule #3: ankyrin
| Macromolecule | Name: ankyrin / type: protein_or_peptide / ID: 3 / Details: two ankyrin in the complex / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MPYSVGFREA DAATSFLRAA RSGNLDKALD HLRNGVDINT CNQNGLNGLH LASKEGHVKM VVELLHKEII LETTTKKGNT ALHIAALAGQ DEVVRELVNY GANVNAQSQK GFTPLYMAAQ ENHLEVVKFL LENGANQNVA TEDGFTPLAV ALQQGHENVV AHLINYGTKG ...String: MPYSVGFREA DAATSFLRAA RSGNLDKALD HLRNGVDINT CNQNGLNGLH LASKEGHVKM VVELLHKEII LETTTKKGNT ALHIAALAGQ DEVVRELVNY GANVNAQSQK GFTPLYMAAQ ENHLEVVKFL LENGANQNVA TEDGFTPLAV ALQQGHENVV AHLINYGTKG KVRLPALHIA ARNDDTRTAA VLLQNDPNPD VLSKTGFTPL HIAAHYENLN VAQLLLNRGA SVNFTPQNGI TPLHIASRRG NVIMVRLLLD RGAQIETKTK DELTPLHCAA RNGHVRISEI LLDHGAPIQA KTKNGLSPIH MAAQGDHLDC VRLLLQYDAE IDDITLDHLT PLHVAAHCGH HRVAKVLLDK GAKPNSRALN GFTPLHIACK KNHVRVMELL LKTGASIDAV TESGLTPLHV ASFMGHLPIV KNLLQRGASP NVSNVKVETP LHMAARAGHT EVAKYLLQNK AKVNAKAKDD QTPLHCAARI GHTNMVKLLL ENNANPNLAT TAGHTPLHIA AREGHVETVL ALLEKEASQA CMTKKGFTPL HVAAKYGKVR VAELLLERDA HPNAAGKNGL TPLHVAVHHN NLDIVKLLLP RGGSPHSPAW NGYTPLHIAA KQNQVEVARS LLQYGGSANA ESVQGVTPLH LAAQEGHAEM VALLLSKQAN GNLGNKSGLT PLHLVAQEGH VPVADVLIKH GVMVDATTRM GYTPLHVASH YGNIKLVKFL LQHQADVNAK TKLGYSPLHQ AAQQGHTDIV TLLLKNGASP NEVSSDGTTP LAIAKRLGYI SVTDVLKVVT DETSFVLVSD KHRMSFPETV DEILDVSEDE GEELISFKAE RRDSRDVDEE KELLDFVPKL DQVVESPAIP RIPCAMPETV VIRSEEQEQA SKEYDEDSLI PSSPATETSD NISPVASPVH TGFLVSFMVD ARGGSMRGSR HNGLRVVIPP RTCAAPTRIT CRLVKPQKLS TPPPLAEEEG LASRIIALGP TGAQFLSPVI VEIPHFASHG RGDRELVVLR SENGSVWKEH RSRYGESYLD QILNGMDEEL GSLEELEKKR VCRIITTDFP LYFVIMSRLC QDYDTIGPEG GSLKSKLVPL VQATFPENAV TKRVKLALQA QPVPDELVTK LLGNQATFSP IVTVEPRRRK FHRPIGLRIP LPPSWTDNPR DSGEGDTTSL RLLCSVIGGT DQAQWEDITG TTKLVYANEC ANFTTNVSAR FWLSDCPRTA EAVNFATLLY KELTAVPYMA KFVIFAKMND PREGRLRCYC MTDDKVDKTL EQHENFVEVA RSRDIEVLEG MSLFAELSGN LVPVKKAAQQ RSFHFQSFRE NRLAMPVKVR DSSREPGGSL SFLRKAMKYE DTQHILCHLN ITMPPCAKGS GAEDRRRTPT PLALRYSILS ESTPGSLSGT EQAEMKMAVI SEHLGLSWAE LARELQFSVE DINRIRVENP NSLLEQSVAL LNLWVIREGQ NANMENLYTA LQSIDRGEIV NMLEGSGRQS RNLKPDRRHT DRDYSLSPSQ MNGYSSLQDE LLSPASLGCA LSSPLRADQY WNEVAVLDAI PLAATEHDTM LEMSDMQVWS AGLTPSLVTA EDSSLECSKA EDSDATGHEW KLEGALSEEP RGPELGSLEL VEDDTVDSDA TNGLIDLLEQ EEGQRSEEKL PGSKRQDDAT GAGQDSENEV SLVSGHQRGQ ARITHSPTVS QVTERSQDRL QDWDADGSIV SYLQDAAQGS WQEEVTQGPH SFQGTSTMTE GLEPGGSQEY EKVLVSVSEH TWTEQPEAES SQADRDRRQQ GQEEQVQEAK NTFTQVVQGN EFQNIPGEQV TEEQFTDEQG NIVTKKIIRK VVRQIDLSSA DAAQEHEEVT VEGPLEDPSE LEVDIDYFMK HSKDHTSTPN P UniProtKB: Ankyrin-1 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2.5 mg/mL | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| |||||||||||||||
| Grid | Model: UltrAuFoil R1.2/1.3 / Material: GOLD / Mesh: 300 / Support film - Material: GOLD / Support film - topology: HOLEY | |||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Temperature | Min: 100.0 K / Max: 100.0 K |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Number grids imaged: 1 / Number real images: 21187 / Average exposure time: 2.0 sec. / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 3.5 µm / Nominal defocus min: 1.8 µm / Nominal magnification: 81000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: AB INITIO MODEL |
|---|
 Movie
Movie Controller
Controller





























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)