[English] 日本語
 Yorodumi
Yorodumi- EMDB-26147: Cryo-EM structure of human band 3-protein 4.2 complex (B2P2vertical) -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of human band 3-protein 4.2 complex (B2P2vertical) | |||||||||
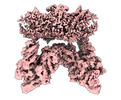
 Map data Map data | band 3-protein 4.2 complex in dimer conformation | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Red blood cell / Ankyrin complex / membrane protein / band 3 / protein 4.2 | |||||||||
| Function / homology |  Function and homology information Function and homology informationhemoglobin metabolic process / protein-glutamine gamma-glutamyltransferase activity / pH elevation / Defective SLC4A1 causes hereditary spherocytosis type 4 (HSP4), distal renal tubular acidosis (dRTA) and dRTA with hemolytic anemia (dRTA-HA) / negative regulation of urine volume / Bicarbonate transporters / intracellular monoatomic ion homeostasis / ankyrin-1 complex / monoatomic anion transmembrane transporter activity / plasma membrane phospholipid scrambling ...hemoglobin metabolic process / protein-glutamine gamma-glutamyltransferase activity / pH elevation / Defective SLC4A1 causes hereditary spherocytosis type 4 (HSP4), distal renal tubular acidosis (dRTA) and dRTA with hemolytic anemia (dRTA-HA) / negative regulation of urine volume / Bicarbonate transporters / intracellular monoatomic ion homeostasis / ankyrin-1 complex / monoatomic anion transmembrane transporter activity / plasma membrane phospholipid scrambling / chloride:bicarbonate antiporter activity / solute:inorganic anion antiporter activity / bicarbonate transport / bicarbonate transmembrane transporter activity / monoatomic anion transport / chloride transport / chloride transmembrane transporter activity / ankyrin binding / hemoglobin binding / erythrocyte maturation / erythrocyte development / negative regulation of glycolytic process through fructose-6-phosphate / cortical cytoskeleton / spleen development / protein-membrane adaptor activity / chloride transmembrane transport / regulation of intracellular pH / protein localization to plasma membrane / Erythrocytes take up oxygen and release carbon dioxide / Erythrocytes take up carbon dioxide and release oxygen / structural constituent of cytoskeleton / multicellular organismal-level iron ion homeostasis / transmembrane transport / cytoplasmic side of plasma membrane / Z disc / cell morphogenesis / blood coagulation / regulation of cell shape / blood microparticle / basolateral plasma membrane / cytoskeleton / protein homodimerization activity / extracellular exosome / ATP binding / membrane / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 4.6 Å | |||||||||
 Authors Authors | Xia X / Liu SH | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2022 Journal: Nat Struct Mol Biol / Year: 2022Title: Structure, dynamics and assembly of the ankyrin complex on human red blood cell membrane. Authors: Xian Xia / Shiheng Liu / Z Hong Zhou /  Abstract: The cytoskeleton of a red blood cell (RBC) is anchored to the cell membrane by the ankyrin complex. This complex is assembled during RBC genesis and comprises primarily band 3, protein 4.2 and ...The cytoskeleton of a red blood cell (RBC) is anchored to the cell membrane by the ankyrin complex. This complex is assembled during RBC genesis and comprises primarily band 3, protein 4.2 and ankyrin, whose mutations contribute to numerous human inherited diseases. High-resolution structures of the ankyrin complex have been long sought-after to understand its assembly and disease-causing mutations. Here, we analyzed native complexes on the human RBC membrane by stepwise fractionation. Cryo-electron microscopy structures of nine band-3-associated complexes reveal that protein 4.2 stabilizes the cytoplasmic domain of band 3 dimer. In turn, the superhelix-shaped ankyrin binds to this protein 4.2 via ankyrin repeats (ARs) 6-13 and to another band 3 dimer via ARs 17-20, bridging two band 3 dimers in the ankyrin complex. Integration of these structures with both prior data and our biochemical data supports a model of ankyrin complex assembly during erythropoiesis and identifies interactions essential for the mechanical stability of RBC. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_26147.map.gz emd_26147.map.gz | 115.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-26147-v30.xml emd-26147-v30.xml emd-26147.xml emd-26147.xml | 16.2 KB 16.2 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_26147.png emd_26147.png | 141.7 KB | ||
| Filedesc metadata |  emd-26147.cif.gz emd-26147.cif.gz | 6.3 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-26147 http://ftp.pdbj.org/pub/emdb/structures/EMD-26147 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26147 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26147 | HTTPS FTP |
-Validation report
| Summary document |  emd_26147_validation.pdf.gz emd_26147_validation.pdf.gz | 616.8 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_26147_full_validation.pdf.gz emd_26147_full_validation.pdf.gz | 616.4 KB | Display | |
| Data in XML |  emd_26147_validation.xml.gz emd_26147_validation.xml.gz | 6.2 KB | Display | |
| Data in CIF |  emd_26147_validation.cif.gz emd_26147_validation.cif.gz | 7 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26147 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26147 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26147 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26147 | HTTPS FTP |
-Related structure data
| Related structure data |  7tw1MC  7tvzC  7tw0C  7tw2C  7tw3C  7tw5C  7tw6C C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_26147.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_26147.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | band 3-protein 4.2 complex in dimer conformation | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.1 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
- Sample components
Sample components
-Entire : High-salt fraction 1 from human red blood cell membrane
| Entire | Name: High-salt fraction 1 from human red blood cell membrane |
|---|---|
| Components |
|
-Supramolecule #1: High-salt fraction 1 from human red blood cell membrane
| Supramolecule | Name: High-salt fraction 1 from human red blood cell membrane type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 350 KDa |
-Macromolecule #1: Band 3 anion transport protein
| Macromolecule | Name: Band 3 anion transport protein / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 101.883859 KDa |
| Sequence | String: MEELQDDYED MMEENLEQEE YEDPDIPESQ MEEPAAHDTE ATATDYHTTS HPGTHKVYVE LQELVMDEKN QELRWMEAAR WVQLEENLG ENGAWGRPHL SHLTFWSLLE LRRVFTKGTV LLDLQETSLA GVANQLLDRF IFEDQIRPQD REELLRALLL K HSHAGELE ...String: MEELQDDYED MMEENLEQEE YEDPDIPESQ MEEPAAHDTE ATATDYHTTS HPGTHKVYVE LQELVMDEKN QELRWMEAAR WVQLEENLG ENGAWGRPHL SHLTFWSLLE LRRVFTKGTV LLDLQETSLA GVANQLLDRF IFEDQIRPQD REELLRALLL K HSHAGELE ALGGVKPAVL TRSGDPSQPL LPQHSSLETQ LFCEQGDGGT EGHSPSGILE KIPPDSEATL VLVGRADFLE QP VLGFVRL QEAAELEAVE LPVPIRFLFV LLGPEAPHID YTQLGRAAAT LMSERVFRID AYMAQSRGEL LHSLEGFLDC SLV LPPTDA PSEQALLSLV PVQRELLRRR YQSSPAKPDS SFYKGLDLNG GPDDPLQQTG QLFGGLVRDI RRRYPYYLSD ITDA FSPQV LAAVIFIYFA ALSPAITFGG LLGEKTRNQM GVSELLISTA VQGILFALLG AQPLLVVGFS GPLLVFEEAF FSFCE TNGL EYIVGRVWIG FWLILLVVLV VAFEGSFLVR FISRYTQEIF SFLISLIFIY ETFSKLIKIF QDHPLQKTYN YNVLMV PKP QGPLPNTALL SLVLMAGTFF FAMMLRKFKN SSYFPGKLRR VIGDFGVPIS ILIMVLVDFF IQDTYTQKLS VPDGFKV SN SSARGWVIHP LGLRSEFPIW MMFASALPAL LVFILIFLES QITTLIVSKP ERKMVKGSGF HLDLLLVVGM GGVAALFG M PWLSATTVRS VTHANALTVM GKASTPGAAA QIQEVKEQRI SGLLVAVLVG LSILMEPILS RIPLAVLFGI FLYMGVTSL SGIQLFDRIL LLFKPPKYHP DVPYVKRVKT WRMHLFTGIQ IICLAVLWVV KSTPASLALP FVLILTVPLR RVLLPLIFRN VELQCLDAD DAKATFDEEE GRDEYDEVAM PV UniProtKB: Band 3 anion transport protein |
-Macromolecule #2: Protein 4.2
| Macromolecule | Name: Protein 4.2 / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 77.096914 KDa |
| Sequence | String: MGQALGIKSC DFQAARNNEE HHTKALSSRR LFVRRGQPFT IILYFRAPVR AFLPALKKVA LTAQTGEQPS KINRTQATFP ISSLGDRKW WSAVVEERDA QSWTISVTTP ADAVIGHYSL LLQVSGRKQL LLGQFTLLFN PWNREDAVFL KNEAQRMEYL L NQNGLIYL ...String: MGQALGIKSC DFQAARNNEE HHTKALSSRR LFVRRGQPFT IILYFRAPVR AFLPALKKVA LTAQTGEQPS KINRTQATFP ISSLGDRKW WSAVVEERDA QSWTISVTTP ADAVIGHYSL LLQVSGRKQL LLGQFTLLFN PWNREDAVFL KNEAQRMEYL L NQNGLIYL GTADCIQAES WDFGQFEGDV IDLSLRLLSK DKQVEKWSQP VHVARVLGAL LHFLKEQRVL PTPQTQATQE GA LLNKRRG SVPILRQWLT GRGRPVYDGQ AWVLAAVACT VLRCLGIPAR VVTTFASAQG TGGRLLIDEY YNEEGLQNGE GQR GRIWIF QTSTECWMTR PALPQGYDGW QILHPSAPNG GGVLGSCDLV PVRAVKEGTL GLTPAVSDLF AAINASCVVW KCCE DGTLE LTDSNTKYVG NNISTKGVGS DRCEDITQNY KYPEGSLQEK EVLERVEKEK MEREKDNGIR PPSLETASPL YLLLK APSS LPLRGDAQIS VTLVNHSEQE KAVQLAIGVQ AVHYNGVLAA KLWRKKLHLT LSANLEKIIT IGLFFSNFER NPPENT FLR LTAMATHSES NLSCFAQEDI AICRPHLAIK MPEKAEQYQP LTASVSLQNS LDAPMEDCVI SILGRGLIHR ERSYRFR SV WPENTMCAKF QFTPTHVGLQ RLTVEVDCNM FQNLTNYKSV TVVAPELSA UniProtKB: Protein 4.2 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2.5 mg/mL | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| |||||||||||||||
| Grid | Model: UltrAuFoil R1.2/1.3 / Material: GOLD / Mesh: 300 / Support film - Material: GOLD / Support film - topology: HOLEY | |||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Temperature | Min: 100.0 K / Max: 100.0 K |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Number grids imaged: 1 / Number real images: 20842 / Average exposure time: 2.0 sec. / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 3.5 µm / Nominal defocus min: 1.8 µm / Nominal magnification: 81000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: AB INITIO MODEL / Overall B value: 212 |
|---|---|
| Output model |  PDB-7tw1: |
 Movie
Movie Controller
Controller















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)




















