[English] 日本語
 Yorodumi
Yorodumi- EMDB-26079: Apo CaKip3[2-436]-L2-mutant(HsKHC) in complex with a microtubule -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Apo CaKip3[2-436]-L2-mutant(HsKHC) in complex with a microtubule | ||||||||||||
 Map data Map data | Primary map, locally refined on the central asymmetric unit | ||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | Kip3 / kinesin / motility / microtubule / tubulin / MOTOR PROTEIN | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationregulation of modification of synapse structure, modulating synaptic transmission / plus-end-directed vesicle transport along microtubule / cytoplasm organization / cytolytic granule membrane / anterograde dendritic transport of neurotransmitter receptor complex / anterograde neuronal dense core vesicle transport / mitocytosis / retrograde neuronal dense core vesicle transport / anterograde axonal protein transport / ciliary rootlet ...regulation of modification of synapse structure, modulating synaptic transmission / plus-end-directed vesicle transport along microtubule / cytoplasm organization / cytolytic granule membrane / anterograde dendritic transport of neurotransmitter receptor complex / anterograde neuronal dense core vesicle transport / mitocytosis / retrograde neuronal dense core vesicle transport / anterograde axonal protein transport / ciliary rootlet / lysosome localization / positive regulation of potassium ion transport / plus-end-directed microtubule motor activity / Kinesins / vesicle transport along microtubule / RHO GTPases activate KTN1 / Microtubule-dependent trafficking of connexons from Golgi to the plasma membrane / Resolution of Sister Chromatid Cohesion / Hedgehog 'off' state / Cilium Assembly / Intraflagellar transport / COPI-dependent Golgi-to-ER retrograde traffic / Mitotic Prometaphase / Carboxyterminal post-translational modifications of tubulin / RHOH GTPase cycle / EML4 and NUDC in mitotic spindle formation / Sealing of the nuclear envelope (NE) by ESCRT-III / Kinesins / PKR-mediated signaling / Separation of Sister Chromatids / The role of GTSE1 in G2/M progression after G2 checkpoint / Aggrephagy / kinesin complex / RHO GTPases activate IQGAPs / RHO GTPases Activate Formins / HSP90 chaperone cycle for steroid hormone receptors (SHR) in the presence of ligand / microtubule motor activity / MHC class II antigen presentation / Recruitment of NuMA to mitotic centrosomes / mitochondrion transport along microtubule / centrosome localization / COPI-dependent Golgi-to-ER retrograde traffic / COPI-mediated anterograde transport / stress granule disassembly / natural killer cell mediated cytotoxicity / microtubule-based movement / Insulin processing / synaptic vesicle transport / postsynaptic cytosol / microtubule-based process / phagocytic vesicle / sperm end piece / axon cytoplasm / MHC class II antigen presentation / dendrite cytoplasm / axon guidance / positive regulation of synaptic transmission, GABAergic / positive regulation of protein localization to plasma membrane / regulation of membrane potential / structural constituent of cytoskeleton / cellular response to type II interferon / microtubule cytoskeleton organization / centriolar satellite / Signaling by ALK fusions and activated point mutants / mitotic cell cycle / microtubule cytoskeleton / nuclear membrane / microtubule binding / vesicle / Hydrolases; Acting on acid anhydrides; Acting on GTP to facilitate cellular and subcellular movement / microtubule / cadherin binding / GTPase activity / GTP binding / protein-containing complex binding / perinuclear region of cytoplasm / ATP hydrolysis activity / mitochondrion / ATP binding / metal ion binding / identical protein binding / membrane / cytoplasm / cytosol Similarity search - Function | ||||||||||||
| Biological species |  Candida albicans (yeast) / Candida albicans (yeast) /  | ||||||||||||
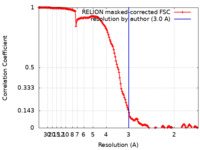
| Method | helical reconstruction / cryo EM / Resolution: 3.0 Å | ||||||||||||
 Authors Authors | Benoit MPMH / Asenjo AB / Hunter B / Allingham JS / Sosa H | ||||||||||||
| Funding support |  United States, United States,  Canada, 3 items Canada, 3 items
| ||||||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2022 Journal: Nat Commun / Year: 2022Title: Kinesin-8-specific loop-2 controls the dual activities of the motor domain according to tubulin protofilament shape. Authors: Byron Hunter / Matthieu P M H Benoit / Ana B Asenjo / Caitlin Doubleday / Daria Trofimova / Corey Frazer / Irsa Shoukat / Hernando Sosa / John S Allingham /   Abstract: Kinesin-8s are dual-activity motor proteins that can move processively on microtubules and depolymerize microtubule plus-ends, but their mechanism of combining these distinct activities remains ...Kinesin-8s are dual-activity motor proteins that can move processively on microtubules and depolymerize microtubule plus-ends, but their mechanism of combining these distinct activities remains unclear. We addressed this by obtaining cryo-EM structures (2.6-3.9 Å) of Candida albicans Kip3 in different catalytic states on the microtubule lattice and on a curved microtubule end mimic. We also determined a crystal structure of microtubule-unbound CaKip3-ADP (2.0 Å) and analyzed the biochemical activity of CaKip3 and kinesin-1 mutants. These data reveal that the microtubule depolymerization activity of kinesin-8 originates from conformational changes of its motor core that are amplified by dynamic contacts between its extended loop-2 and tubulin. On curved microtubule ends, loop-1 inserts into preceding motor domains, forming head-to-tail arrays of kinesin-8s that complement loop-2 contacts with curved tubulin and assist depolymerization. On straight tubulin protofilaments in the microtubule lattice, loop-2-tubulin contacts inhibit conformational changes in the motor core, but in the ADP-Pi state these contacts are relaxed, allowing neck-linker docking for motility. We propose that these tubulin shape-induced alternations between pro-microtubule-depolymerization and pro-motility kinesin states, regulated by loop-2, are the key to the dual activity of kinesin-8 motors. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_26079.map.gz emd_26079.map.gz | 39.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-26079-v30.xml emd-26079-v30.xml emd-26079.xml emd-26079.xml | 23.5 KB 23.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_26079_fsc.xml emd_26079_fsc.xml | 14.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_26079.png emd_26079.png | 128 KB | ||
| Masks |  emd_26079_msk_1.map emd_26079_msk_1.map emd_26079_msk_2.map emd_26079_msk_2.map emd_26079_msk_3.map emd_26079_msk_3.map emd_26079_msk_4.map emd_26079_msk_4.map | 274.6 MB 274.6 MB 274.6 MB 274.6 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-26079.cif.gz emd-26079.cif.gz | 7.9 KB | ||
| Others |  emd_26079_half_map_1.map.gz emd_26079_half_map_1.map.gz emd_26079_half_map_2.map.gz emd_26079_half_map_2.map.gz | 218.6 MB 218.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-26079 http://ftp.pdbj.org/pub/emdb/structures/EMD-26079 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26079 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26079 | HTTPS FTP |
-Related structure data
| Related structure data |  7tr2MC  7lffC  7tqxC  7tqyC  7tqzC  7tr0C  7tr1C  7tr3C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_26079.map.gz / Format: CCP4 / Size: 274.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_26079.map.gz / Format: CCP4 / Size: 274.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Primary map, locally refined on the central asymmetric unit | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.849 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_26079_msk_1.map emd_26079_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Mask #2
| File |  emd_26079_msk_2.map emd_26079_msk_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Mask #3
| File |  emd_26079_msk_3.map emd_26079_msk_3.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Mask #4
| File |  emd_26079_msk_4.map emd_26079_msk_4.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map from the gold Standard refinement
| File | emd_26079_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map from the gold Standard refinement | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map from the gold Standard refinement
| File | emd_26079_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map from the gold Standard refinement | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
+Entire : Apo CaKip3[2-436]-L2-mutant(HsKHC) in complex with a microtubule
+Supramolecule #1: Apo CaKip3[2-436]-L2-mutant(HsKHC) in complex with a microtubule
+Supramolecule #2: CaKip3[2-436] with L2 swapped with the one of HsKHC, bound to AMP-PNP
+Supramolecule #3: 15R microtubule
+Macromolecule #1: Kinesin-like protein,Kinesin-1 heavy chain
+Macromolecule #2: Tubulin alpha-1B chain
+Macromolecule #3: Tubulin beta-2B chain
+Macromolecule #4: GUANOSINE-5'-TRIPHOSPHATE
+Macromolecule #5: MAGNESIUM ION
+Macromolecule #6: GUANOSINE-5'-DIPHOSPHATE
+Macromolecule #7: TAXOL
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | filament |
- Sample preparation
Sample preparation
| Buffer | pH: 6.8 Component:
| ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Grid | Model: UltrAuFoil R2/2 / Support film - Material: GOLD / Support film - topology: HOLEY / Pretreatment - Type: PLASMA CLEANING | ||||||||||
| Vitrification | Cryogen name: ETHANE / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Digitization - Frames/image: 1-40 / Average electron dose: 64.6 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.5 µm / Nominal defocus min: 0.7000000000000001 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT |
|---|---|
| Output model |  PDB-7tr2: |
 Movie
Movie Controller
Controller



























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)