+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | CryoEM structure of JetD from Pseudomonas aeruginosa | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Wadjet / Bacterial defense systems / JetD / Anti-plasmid defense system / EptD / MksG / Toprim domain / Nuclease / Topoisomerase / DNA BINDING PROTEIN | |||||||||
| Function / homology | Uncharacterised conserved protein UCP028408 / Wadjet protein JetD, C-terminal / Domain of unknown function DUF3322 / Wadjet protein JetD, C-terminal / Uncharacterized protein conserved in bacteria N-term (DUF3322) / DUF3322 and DUF2220 domain-containing protein Function and homology information Function and homology information | |||||||||
| Biological species |  Pseudomonas aeruginosa PA14 (bacteria) Pseudomonas aeruginosa PA14 (bacteria) | |||||||||
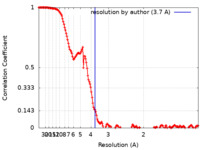
| Method | single particle reconstruction / cryo EM / Resolution: 3.7 Å | |||||||||
 Authors Authors | Deep A / Gu Y | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Mol Cell / Year: 2022 Journal: Mol Cell / Year: 2022Title: The SMC-family Wadjet complex protects bacteria from plasmid transformation by recognition and cleavage of closed-circular DNA. Authors: Amar Deep / Yajie Gu / Yong-Qi Gao / Kaori M Ego / Mark A Herzik / Huilin Zhou / Kevin D Corbett /  Abstract: Self versus non-self discrimination is a key element of innate and adaptive immunity across life. In bacteria, CRISPR-Cas and restriction-modification systems recognize non-self nucleic acids through ...Self versus non-self discrimination is a key element of innate and adaptive immunity across life. In bacteria, CRISPR-Cas and restriction-modification systems recognize non-self nucleic acids through their sequence and their methylation state, respectively. Here, we show that the Wadjet defense system recognizes DNA topology to protect its host against plasmid transformation. By combining cryoelectron microscopy with cross-linking mass spectrometry, we show that Wadjet forms a complex similar to the bacterial condensin complex MukBEF, with a novel nuclease subunit similar to a type II DNA topoisomerase. Wadjet specifically cleaves closed-circular DNA in a reaction requiring ATP hydrolysis by the structural maintenance of chromosome (SMC) ATPase subunit JetC, suggesting that the complex could use DNA loop extrusion to sense its substrate's topology, then specifically activate the nuclease subunit JetD to cleave plasmid DNA. Overall, our data reveal how bacteria have co-opted a DNA maintenance machine to specifically recognize and destroy foreign DNAs through topology sensing. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_25913.map.gz emd_25913.map.gz | 169.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-25913-v30.xml emd-25913-v30.xml emd-25913.xml emd-25913.xml | 28 KB 28 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_25913_fsc.xml emd_25913_fsc.xml | 15.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_25913.png emd_25913.png | 104.8 KB | ||
| Filedesc metadata |  emd-25913.cif.gz emd-25913.cif.gz | 6.1 KB | ||
| Others |  emd_25913_additional_1.map.gz emd_25913_additional_1.map.gz emd_25913_additional_2.map.gz emd_25913_additional_2.map.gz emd_25913_additional_3.map.gz emd_25913_additional_3.map.gz emd_25913_additional_4.map.gz emd_25913_additional_4.map.gz emd_25913_additional_5.map.gz emd_25913_additional_5.map.gz emd_25913_additional_6.map.gz emd_25913_additional_6.map.gz emd_25913_half_map_1.map.gz emd_25913_half_map_1.map.gz emd_25913_half_map_2.map.gz emd_25913_half_map_2.map.gz | 318.3 MB 318.3 MB 170.2 MB 318.1 MB 318.1 MB 170 MB 318.7 MB 318.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-25913 http://ftp.pdbj.org/pub/emdb/structures/EMD-25913 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-25913 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-25913 | HTTPS FTP |
-Validation report
| Summary document |  emd_25913_validation.pdf.gz emd_25913_validation.pdf.gz | 1 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_25913_full_validation.pdf.gz emd_25913_full_validation.pdf.gz | 1 MB | Display | |
| Data in XML |  emd_25913_validation.xml.gz emd_25913_validation.xml.gz | 24.1 KB | Display | |
| Data in CIF |  emd_25913_validation.cif.gz emd_25913_validation.cif.gz | 31.3 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-25913 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-25913 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-25913 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-25913 | HTTPS FTP |
-Related structure data
| Related structure data |  7tilMC  8dk1C  8dk2C  8dk3C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_25913.map.gz / Format: CCP4 / Size: 343 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_25913.map.gz / Format: CCP4 / Size: 343 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.65 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: #6
| File | emd_25913_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: #5
| File | emd_25913_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: #4
| File | emd_25913_additional_3.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: #3
| File | emd_25913_additional_4.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: #2
| File | emd_25913_additional_5.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: #1
| File | emd_25913_additional_6.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_25913_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_25913_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : JetD
| Entire | Name: JetD |
|---|---|
| Components |
|
-Supramolecule #1: JetD
| Supramolecule | Name: JetD / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all / Details: A Pseudomonas aeruginosa protein. |
|---|---|
| Source (natural) | Organism:  Pseudomonas aeruginosa PA14 (bacteria) Pseudomonas aeruginosa PA14 (bacteria) |
| Molecular weight | Theoretical: 85.6 kDa/nm |
-Macromolecule #1: JetD
| Macromolecule | Name: JetD / type: protein_or_peptide / ID: 1 Details: Pseudomonas aeruginosa PA14 JetD protein with N-terminal TEV cleavable 6XHis tag. Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Pseudomonas aeruginosa PA14 (bacteria) / Strain: UCBPP-PA14 Pseudomonas aeruginosa PA14 (bacteria) / Strain: UCBPP-PA14 |
| Molecular weight | Theoretical: 45.132164 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MKSSHHHHHH ENLYFQSNAK SPTDIGRNLA RQWQRSSVRL ERLLNPGSWP QSLNIGKPSA RLFTDNIQAV LRHVENWRSV NVGKVDWEP VSYRAGLAPI SLPMRWHLRS PSEWIAASDD PRVSQEYAQL EYLVEQVDSA FHTLLVAQRS LWLTRTPEEV V ATAQLATQ ...String: MKSSHHHHHH ENLYFQSNAK SPTDIGRNLA RQWQRSSVRL ERLLNPGSWP QSLNIGKPSA RLFTDNIQAV LRHVENWRSV NVGKVDWEP VSYRAGLAPI SLPMRWHLRS PSEWIAASDD PRVSQEYAQL EYLVEQVDSA FHTLLVAQRS LWLTRTPEEV V ATAQLATQ LAPGCAQGRP LRLLAEHGVD TKFFERNATL LTKLLDERFE GAASEQGLTT FLDAFEESSH WVLVVPLQPG LL PFKRLRL TTSELAETPL PGSRLLVVEN EQCVHLLPET LPDTLAVLGA GLDLHWLASA HLAGKQIGYW GDMDTWGLLM LAR ARLHQP AVEALLMEQE LFEQHSQGNT VVEPAKALES APPGLLADEA DFYRYLLVQE RGRLEQEYLP KAQVELAIRK WAR UniProtKB: DUF3322 and DUF2220 domain-containing protein |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1.10 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
Details: Prepared using deionized water and filtered strelized. | ||||||||||||
| Grid | Model: UltrAuFoil R1.2/1.3 / Material: GOLD / Mesh: 300 / Support film - Material: GOLD / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 12 sec. | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 QUANTUM (4k x 4k) / Average electron dose: 62.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: AB INITIO MODEL |
|---|---|
| Output model |  PDB-7til: |
 Movie
Movie Controller
Controller







 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)