[English] 日本語
 Yorodumi
Yorodumi- EMDB-27482: CryoEM structure of Pseudomonas aeruginosa PA14 JetC ATPase domai... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | CryoEM structure of Pseudomonas aeruginosa PA14 JetC ATPase domain bound to DNA and cWHD domain of JetA | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Wadjet / Bacterial defense systems / JetA / JetC / Anti-plasmid defense system / EptA / EptC / MksB / MksF / SMC / DNA BINDING PROTEIN-DNA complex | |||||||||
| Function / homology | Protein of unknown function DUF3375 / Protein of unknown function (DUF3375) / P-loop containing nucleoside triphosphate hydrolase / DUF3375 domain-containing protein / ATP synthase Function and homology information Function and homology information | |||||||||
| Biological species |  Pseudomonas aeruginosa PA14 (bacteria) / synthetic construct (others) Pseudomonas aeruginosa PA14 (bacteria) / synthetic construct (others) | |||||||||
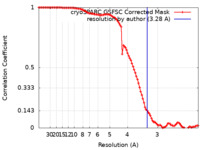
| Method | single particle reconstruction / cryo EM / Resolution: 3.28 Å | |||||||||
 Authors Authors | Deep A / Gu Y / Gao Y / Ego K / Herzik M / Zhou H / Corbett K | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Mol Cell / Year: 2022 Journal: Mol Cell / Year: 2022Title: The SMC-family Wadjet complex protects bacteria from plasmid transformation by recognition and cleavage of closed-circular DNA. Authors: Amar Deep / Yajie Gu / Yong-Qi Gao / Kaori M Ego / Mark A Herzik / Huilin Zhou / Kevin D Corbett /  Abstract: Self versus non-self discrimination is a key element of innate and adaptive immunity across life. In bacteria, CRISPR-Cas and restriction-modification systems recognize non-self nucleic acids through ...Self versus non-self discrimination is a key element of innate and adaptive immunity across life. In bacteria, CRISPR-Cas and restriction-modification systems recognize non-self nucleic acids through their sequence and their methylation state, respectively. Here, we show that the Wadjet defense system recognizes DNA topology to protect its host against plasmid transformation. By combining cryoelectron microscopy with cross-linking mass spectrometry, we show that Wadjet forms a complex similar to the bacterial condensin complex MukBEF, with a novel nuclease subunit similar to a type II DNA topoisomerase. Wadjet specifically cleaves closed-circular DNA in a reaction requiring ATP hydrolysis by the structural maintenance of chromosome (SMC) ATPase subunit JetC, suggesting that the complex could use DNA loop extrusion to sense its substrate's topology, then specifically activate the nuclease subunit JetD to cleave plasmid DNA. Overall, our data reveal how bacteria have co-opted a DNA maintenance machine to specifically recognize and destroy foreign DNAs through topology sensing. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_27482.map.gz emd_27482.map.gz | 51.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-27482-v30.xml emd-27482-v30.xml emd-27482.xml emd-27482.xml | 26.4 KB 26.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_27482_fsc.xml emd_27482_fsc.xml | 9.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_27482.png emd_27482.png | 98.1 KB | ||
| Filedesc metadata |  emd-27482.cif.gz emd-27482.cif.gz | 8.3 KB | ||
| Others |  emd_27482_half_map_1.map.gz emd_27482_half_map_1.map.gz emd_27482_half_map_2.map.gz emd_27482_half_map_2.map.gz | 95.6 MB 95.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-27482 http://ftp.pdbj.org/pub/emdb/structures/EMD-27482 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27482 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27482 | HTTPS FTP |
-Related structure data
| Related structure data |  8dk3MC  7tilC  8dk1C  8dk2C C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_27482.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_27482.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.1 Å | ||||||||||||||||||||||||||||||||||||
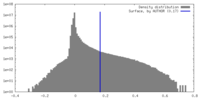
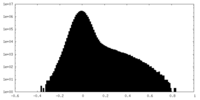
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_27482_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
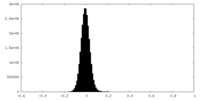
| Density Histograms |
-Half map: #2
| File | emd_27482_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
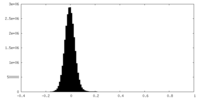
| Density Histograms |
- Sample components
Sample components
-Entire : JetC head ATPase domain in complex with DNA, and cWHD of JetA
| Entire | Name: JetC head ATPase domain in complex with DNA, and cWHD of JetA |
|---|---|
| Components |
|
-Supramolecule #1: JetC head ATPase domain in complex with DNA, and cWHD of JetA
| Supramolecule | Name: JetC head ATPase domain in complex with DNA, and cWHD of JetA type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#4 |
|---|---|
| Source (natural) | Organism:  Pseudomonas aeruginosa PA14 (bacteria) Pseudomonas aeruginosa PA14 (bacteria) |
| Molecular weight | Theoretical: 110 KDa |
-Macromolecule #1: JetC
| Macromolecule | Name: JetC / type: protein_or_peptide / ID: 1 / Details: The sequence contains mutation: E1022Q. / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Pseudomonas aeruginosa PA14 (bacteria) Pseudomonas aeruginosa PA14 (bacteria) |
| Molecular weight | Theoretical: 126.68318 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MKSSHHHHHH ENLYFQSNAK QALLWRDSET FILTSIELYN WGGFQGYHRA EIDPSGTAVI GPTGSGKTTL VDALMTLLCA NPRYNLAST GGHESDRDLV SYVRGVTGPG DGGVEQSHIA RQGKTVTAIA ATLERDGAQV RLGAVLWFEG TSSSASDLKK L WLLSESPE ...String: MKSSHHHHHH ENLYFQSNAK QALLWRDSET FILTSIELYN WGGFQGYHRA EIDPSGTAVI GPTGSGKTTL VDALMTLLCA NPRYNLAST GGHESDRDLV SYVRGVTGPG DGGVEQSHIA RQGKTVTAIA ATLERDGAQV RLGAVLWFEG TSSSASDLKK L WLLSESPE QTLEHWLSQH HAGGMRALRQ MEKDGMGIWP YPSKKAFLAR LRDYFEVGEN AFTLLNRAAG LKQLNSIDEI FR ELVLDDR SAFERAAEVA SSFDDLTDIH RELETARKQQ RSLQPVADGW ERYRALQEQL QDKQALEGIL PVWFAEQGYR LWL AETNRL EKEHKQAELD QAQCRSQLEI QKGVVDQHRQ RYLRVGGAGI DQLRGRIADW VRECDKRRLK AEQYQRLAKG LGLA DELSA AALEENQQQI AARLEILAQQ TTDARQKAFD AGLVQQELNG RLQSLQQERA EVERRPGSNL PGHFHAFRGD LAQEL GVDE SALPFVAELV QVKPEELAWR GAIERAIGSH RLRILVPQGS SQAALRWVNQ RHNRLHVRLL EVKEPSSRPV FFDDGF TRK LTFKEHPYRE AVKALLADND RHCVESTEQL RHTPHAMTAQ GLMSGKERFF DKQDQKRLDE DWLTGFDNRD RLAFLAE QI REVNEQLVPA KLALDAAQGD VGQLESQASL LQRIEELQFD DIDRPGAERQ LQSLRTQLDT LTRPDSNLAV IKAELDQA E ALRESLDQQL QRLIEQCVQL KTQFDQAASA TRKAYRGAEK GLSDTQRELA QAHFPILSTD DLGDIDELER KHTRELQGQ LKTLGEKLGD QKTELAKRMS DALKADTGAL AEVGRELVDV PRYLERLRVL TEEALPEKLK RFLEYLNRSS DDGVTQLLSY IDHEVSMIE ERLDDLNSTM QRVDFQPGRY LRLVAKKVIH ESLRTLQHAQ RQLNSARFID DEGESHYKAL QALVGLLKDA C EHSRNQGA KALLDPRFRL EFAVSVIDRE GNNLIETRTG SQGGSGGEKE IIASYVLTAS LSYALCPDGS SRPLFGTIVL DQ AFSRSSH AVAGRIIAAL REFGLHAVFI TPNKEMRLLR HHTRSAVVVH RRGVESSLVS LSWEALDEHH QQRIRAMHEV AH UniProtKB: ATP synthase |
-Macromolecule #2: JetA
| Macromolecule | Name: JetA / type: protein_or_peptide / ID: 2 Details: cWHD domain of JetA observed in complex with dimeric JetC. Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Pseudomonas aeruginosa PA14 (bacteria) Pseudomonas aeruginosa PA14 (bacteria) |
| Molecular weight | Theoretical: 59.2335 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MKSSHHHHHH ENLYFQSNAE ESAQQRSERY VSARSQHPAW LLLASRRAPL VLGCLRTLFE RAHDGIPMED ALQALSEMLA AYASQELYE IDPDATHLQA GRELREWIKR RLVVEREGRI YATDALESAI QFVDSLDSRI MTSTASRLSV VQREIENLET G LNPSPTGR ...String: MKSSHHHHHH ENLYFQSNAE ESAQQRSERY VSARSQHPAW LLLASRRAPL VLGCLRTLFE RAHDGIPMED ALQALSEMLA AYASQELYE IDPDATHLQA GRELREWIKR RLVVEREGRI YATDALESAI QFVDSLDSRI MTSTASRLSV VQREIENLET G LNPSPTGR IASLRRRIQD LEHELARVEA GHVDVLDEAQ AIEGMREVYN LATSLRADFR RVEDSWREAD RALRHSIISE QS HRGEIVD RLLDGQDALL NTPEGRVFES FQQQLRQSAE LEVMRERLRT ILRHPAVPKA LNRPQQRELR WLALRLVRES QAV LQARAR SERDVRGFMK TGLAAEHHRV GQLLNDFFNL ALSVDWQRQS ERRKPACLPP VGVAITGVPA IERLRFKTLD DDDA GELDL SLKPAGLEQI DDDFWDAFDG LDREALIHDT LAVLVEQGRP VSLGELASLL PPAHDLETFA LWLAMAREAG IEVLT EERQ FVELVDEDEQ RWGFNLPYVG LDHEALKDID WEL UniProtKB: DUF3375 domain-containing protein |
-Macromolecule #3: DNA (26-MER)
| Macromolecule | Name: DNA (26-MER) / type: dna / ID: 3 Details: Below mentioned hybridized oligonucleotides were used in the sample preparation. Considering the DNA binding is purely based on the electrostatic charge: polyA/polyT chain was used to ...Details: Below mentioned hybridized oligonucleotides were used in the sample preparation. Considering the DNA binding is purely based on the electrostatic charge: polyA/polyT chain was used to generate the model. JetABC_cryo_Fwd: GTGATAGTTAGAAACGTAATTGACTATAAAGATGATGACGATAAGTAGGATGTCTATAGACCAGG JetABC_cryo_Rev: CCTGGTCTATAGACATCCTACTTATCGTCATCATCTTTATAGTCAATTACGTTTCTAACTATCAC Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 7.864056 KDa |
| Sequence | String: (DT)(DT)(DT)(DT)(DT)(DT)(DT)(DT)(DT)(DT) (DT)(DT)(DT)(DT)(DT)(DT)(DT)(DT)(DT)(DT) (DT)(DT)(DT)(DT)(DT)(DT) |
-Macromolecule #4: DNA (26-MER)
| Macromolecule | Name: DNA (26-MER) / type: dna / ID: 4 Details: Below mentioned hybridized oligonucleotides were used in the sample preparation. Considering the DNA binding is purely based on the electrostatic charge: polyA/polyT chain was used to ...Details: Below mentioned hybridized oligonucleotides were used in the sample preparation. Considering the DNA binding is purely based on the electrostatic charge: polyA/polyT chain was used to generate the model. JetABC_cryo_Fwd: GTGATAGTTAGAAACGTAATTGACTATAAAGATGATGACGATAAGTAGGATGTCTATAGACCAGG JetABC_cryo_Rev: CCTGGTCTATAGACATCCTACTTATCGTCATCATCTTTATAGTCAATTACGTTTCTAACTATCAC Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 8.098421 KDa |
| Sequence | String: (DA)(DA)(DA)(DA)(DA)(DA)(DA)(DA)(DA)(DA) (DA)(DA)(DA)(DA)(DA)(DA)(DA)(DA)(DA)(DA) (DA)(DA)(DA)(DA)(DA)(DA) |
-Macromolecule #5: PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER
| Macromolecule | Name: PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER / type: ligand / ID: 5 / Number of copies: 2 / Formula: AGS |
|---|---|
| Molecular weight | Theoretical: 523.247 Da |
| Chemical component information |  ChemComp-AGS: |
-Macromolecule #6: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 6 / Number of copies: 2 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1.5 mg/mL | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
Details: Prepared using deionized water and filtered sterilized. | ||||||||||||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY | ||||||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 QUANTUM (4k x 4k) / Detector mode: COUNTING / Average electron dose: 50.1 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.5 µm / Nominal magnification: 130000 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: AB INITIO MODEL |
|---|---|
| Output model |  PDB-8dk3: |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)