+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-25898 | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
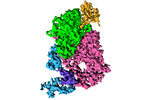
| Title | SARS-CoV-2 nsp12/7/8 complex with a native N-terminus nsp9 | |||||||||||||||
 Map data Map data | SARS-CoV-2 nsp12/7/8 complex with a native N-terminus nsp9 | |||||||||||||||
 Sample Sample |
| |||||||||||||||
 Keywords Keywords | polymerase / NiRAN / capping / nsp9 / VIRAL PROTEIN | |||||||||||||||
| Function / homology |  Function and homology information Function and homology informationprotein guanylyltransferase activity / RNA endonuclease activity producing 3'-phosphomonoesters, hydrolytic mechanism / mRNA guanylyltransferase activity / 5'-3' RNA helicase activity / Lyases; Phosphorus-oxygen lyases / Assembly of the SARS-CoV-2 Replication-Transcription Complex (RTC) / symbiont-mediated suppression of host cytoplasmic pattern recognition receptor signaling pathway via inhibition of TBK1 activity / Maturation of replicase proteins / TRAF3-dependent IRF activation pathway / ISG15-specific peptidase activity ...protein guanylyltransferase activity / RNA endonuclease activity producing 3'-phosphomonoesters, hydrolytic mechanism / mRNA guanylyltransferase activity / 5'-3' RNA helicase activity / Lyases; Phosphorus-oxygen lyases / Assembly of the SARS-CoV-2 Replication-Transcription Complex (RTC) / symbiont-mediated suppression of host cytoplasmic pattern recognition receptor signaling pathway via inhibition of TBK1 activity / Maturation of replicase proteins / TRAF3-dependent IRF activation pathway / ISG15-specific peptidase activity / Transcription of SARS-CoV-2 sgRNAs / snRNP Assembly / Translation of Replicase and Assembly of the Replication Transcription Complex / Replication of the SARS-CoV-2 genome / Hydrolases; Acting on ester bonds; Exoribonucleases producing 5'-phosphomonoesters / double membrane vesicle viral factory outer membrane / SARS coronavirus main proteinase / host cell endoplasmic reticulum-Golgi intermediate compartment / 5'-3' DNA helicase activity / 3'-5'-RNA exonuclease activity / symbiont-mediated degradation of host mRNA / host cell endosome / mRNA guanylyltransferase / symbiont-mediated suppression of host ISG15-protein conjugation / symbiont-mediated suppression of host toll-like receptor signaling pathway / G-quadruplex RNA binding / symbiont-mediated suppression of host cytoplasmic pattern recognition receptor signaling pathway via inhibition of IRF3 activity / omega peptidase activity / mRNA (guanine-N7)-methyltransferase / methyltransferase cap1 / SARS-CoV-2 modulates host translation machinery / host cell Golgi apparatus / symbiont-mediated suppression of host NF-kappaB cascade / DNA helicase / symbiont-mediated perturbation of host ubiquitin-like protein modification / methyltransferase cap1 activity / ubiquitinyl hydrolase 1 / cysteine-type deubiquitinase activity / mRNA 5'-cap (guanine-N7-)-methyltransferase activity / Hydrolases; Acting on peptide bonds (peptidases); Cysteine endopeptidases / single-stranded RNA binding / regulation of autophagy / lyase activity / viral protein processing / host cell perinuclear region of cytoplasm / host cell endoplasmic reticulum membrane / RNA helicase / symbiont-mediated suppression of host type I interferon-mediated signaling pathway / copper ion binding / symbiont-mediated suppression of host gene expression / symbiont-mediated activation of host autophagy / viral translational frameshifting / RNA-directed RNA polymerase / cysteine-type endopeptidase activity / viral RNA genome replication / RNA-directed RNA polymerase activity / DNA-templated transcription / lipid binding / host cell nucleus / SARS-CoV-2 activates/modulates innate and adaptive immune responses / ATP hydrolysis activity / proteolysis / RNA binding / zinc ion binding / ATP binding / membrane Similarity search - Function | |||||||||||||||
| Biological species |  | |||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.18 Å | |||||||||||||||
 Authors Authors | Osinski A / Tagliabracci VS | |||||||||||||||
| Funding support |  United States, 4 items United States, 4 items
| |||||||||||||||
 Citation Citation | Journal: Res Sq / Year: 2022 Title: The mechanism of RNA capping by SARS-CoV-2. Authors: Gina J Park / Adam Osinski / Genaro Hernandez / Jennifer L Eitson / Abir Majumdar / Marco Tonelli / Katie Henzler-Wildman / Krzysztof Pawłowski / Zhe Chen / Yang Li / John W Schoggins / ...Authors: Gina J Park / Adam Osinski / Genaro Hernandez / Jennifer L Eitson / Abir Majumdar / Marco Tonelli / Katie Henzler-Wildman / Krzysztof Pawłowski / Zhe Chen / Yang Li / John W Schoggins / Vincent S Tagliabracci /   Abstract: The SARS-CoV-2 RNA genome contains a 5'-cap that facilitates translation of viral proteins, protection from exonucleases and evasion of the host immune response1-4. How this cap is made is not ...The SARS-CoV-2 RNA genome contains a 5'-cap that facilitates translation of viral proteins, protection from exonucleases and evasion of the host immune response1-4. How this cap is made is not completely understood. Here, we reconstitute the SARS-CoV-2 7MeGpppA2'-O-Me-RNA cap using virally encoded non-structural proteins (nsps). We show that the kinase-like NiRAN domain5 of nsp12 transfers RNA to the amino terminus of nsp9, forming a covalent RNA-protein intermediate (a process termed RNAylation). Subsequently, the NiRAN domain transfers RNA to GDP, forming the cap core structure GpppA-RNA. The nsp146 and nsp167 methyltransferases then add methyl groups to form functional cap structures. Structural analyses of the replication-transcription complex bound to nsp9 identified key interactions that mediate the capping reaction. Furthermore, we demonstrate in a reverse genetics system8 that the N-terminus of nsp9 and the kinase-like active site residues in the NiRAN domain are required for successful SARS-CoV-2 replication. Collectively, our results reveal an unconventional mechanism by which SARS-CoV-2 caps its RNA genome, thus exposing a new target in the development of antivirals to treat COVID-19. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_25898.map.gz emd_25898.map.gz | 118.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-25898-v30.xml emd-25898-v30.xml emd-25898.xml emd-25898.xml | 20.6 KB 20.6 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_25898.png emd_25898.png | 73.9 KB | ||
| Filedesc metadata |  emd-25898.cif.gz emd-25898.cif.gz | 7.2 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-25898 http://ftp.pdbj.org/pub/emdb/structures/EMD-25898 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-25898 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-25898 | HTTPS FTP |
-Related structure data
| Related structure data |  7thmMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_25898.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_25898.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | SARS-CoV-2 nsp12/7/8 complex with a native N-terminus nsp9 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.094 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : SARS-CoV-2 replication-transcription complex with a native N-term...
| Entire | Name: SARS-CoV-2 replication-transcription complex with a native N-terminus nsp9 bound to NiRAN domain of nsp12. |
|---|---|
| Components |
|
-Supramolecule #1: SARS-CoV-2 replication-transcription complex with a native N-term...
| Supramolecule | Name: SARS-CoV-2 replication-transcription complex with a native N-terminus nsp9 bound to NiRAN domain of nsp12. type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#4 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 172.03949 KDa |
-Macromolecule #1: RNA-directed RNA polymerase
| Macromolecule | Name: RNA-directed RNA polymerase / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO / EC number: RNA-directed RNA polymerase |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 106.780977 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: SADAQSFLNR VCGVSAARLT PCGTGTSTDV VYRAFDIYND KVAGFAKFLK TNCCRFQEKD EDDNLIDSYF VVKRHTFSNY QHEETIYNL LKDCPAVAKH DFFKFRIDGD MVPHISRQRL TKYTMADLVY ALRHFDEGNC DTLKEILVTY NCCDDDYFNK K DWYDFVEN ...String: SADAQSFLNR VCGVSAARLT PCGTGTSTDV VYRAFDIYND KVAGFAKFLK TNCCRFQEKD EDDNLIDSYF VVKRHTFSNY QHEETIYNL LKDCPAVAKH DFFKFRIDGD MVPHISRQRL TKYTMADLVY ALRHFDEGNC DTLKEILVTY NCCDDDYFNK K DWYDFVEN PDILRVYANL GERVRQALLK TVQFCDAMRN AGIVGVLTLD NQDLNGNWYD FGDFIQTTPG SGVPVVDSYY SL LMPILTL TRALTAESHV DTDLTKPYIK WDLLKYDFTE ERLKLFDRYF KYWDQTYHPN CVNCLDDRCI LHCANFNVLF STV FPPTSF GPLVRKIFVD GVPFVVSTGY HFRELGVVHN QDVNLHSSRL SFKELLVYAA DPAMHAASGN LLLDKRTTCF SVAA LTNNV AFQTVKPGNF NKDFYDFAVS KGFFKEGSSV ELKHFFFAQD GNAAISDYDY YRYNLPTMCD IRQLLFVVEV VDKYF DCYD GGCINANQVI VNNLDKSAGF PFNKWGKARL YYDSMSYEDQ DALFAYTKRN VIPTITQMNL KYAISAKNRA RTVAGV SIC STMTNRQFHQ KLLKSIAATR GATVVIGTSK FYGGWHNMLK TVYSDVENPH LMGWDYPKCD RAMPNMLRIM ASLVLAR KH TTCCSLSHRF YRLANECAQV LSEMVMCGGS LYVKPGGTSS GDATTAYANS VFNICQAVTA NVNALLSTDG NKIADKYV R NLQHRLYECL YRNRDVDTDF VNEFYAYLRK HFSMMILSDD AVVCFNSTYA SQGLVASIKN FKSVLYYQNN VFMSEAKCW TETDLTKGPH EFCSQHTMLV KQGDDYVYLP YPDPSRILGA GCFVDDIVKT DGTLMIERFV SLAIDAYPLT KHPNQEYADV FHLYLQYIR KLHDELTGHM LDMYSVMLTN DNTSRYWEPE FYEAMYTPHT VLQ UniProtKB: Replicase polyprotein 1ab |
-Macromolecule #2: Non-structural protein 8
| Macromolecule | Name: Non-structural protein 8 / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 21.903047 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: AIASEFSSLP SYAAFATAQE AYEQAVANGD SEVVLKKLKK SLNVAKSEFD RDAAMQRKLE KMADQAMTQM YKQARSEDKR AKVTSAMQT MLFTMLRKLD NDALNNIINN ARDGCVPLNI IPLTTAAKLM VVIPDYNTYK NTCDGTTFTY ASALWEIQQV V DADSKIVQ ...String: AIASEFSSLP SYAAFATAQE AYEQAVANGD SEVVLKKLKK SLNVAKSEFD RDAAMQRKLE KMADQAMTQM YKQARSEDKR AKVTSAMQT MLFTMLRKLD NDALNNIINN ARDGCVPLNI IPLTTAAKLM VVIPDYNTYK NTCDGTTFTY ASALWEIQQV V DADSKIVQ LSEISMDNSP NLAWPLIVTA LRANSAVKLQ UniProtKB: Replicase polyprotein 1ab |
-Macromolecule #3: Non-structural protein 7
| Macromolecule | Name: Non-structural protein 7 / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 9.248804 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: SKMSDVKCTS VVLLSVLQQL RVESSSKLWA QCVQLHNDIL LAKDTTEAFE KMVSLLSVLL SMQGAVDINK LCEEMLDNRA TLQ UniProtKB: Replicase polyprotein 1ab |
-Macromolecule #4: Non-structural protein 9
| Macromolecule | Name: Non-structural protein 9 / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 12.391171 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: NNELSPVALR QMSCAAGTTQ TACTDDNALA YYNTTKGGRF VLALLSDLQD LKWARFPKSD GTGTIYTELE PPCRFVTDTP KGPKVKYLY FIKGLNNLNR GMVLGSLAAT VRLQ UniProtKB: Replicase polyprotein 1ab |
-Macromolecule #5: ZINC ION
| Macromolecule | Name: ZINC ION / type: ligand / ID: 5 / Number of copies: 2 / Formula: ZN |
|---|---|
| Molecular weight | Theoretical: 65.409 Da |
-Macromolecule #6: MANGANESE (II) ION
| Macromolecule | Name: MANGANESE (II) ION / type: ligand / ID: 6 / Number of copies: 1 / Formula: MN |
|---|---|
| Molecular weight | Theoretical: 54.938 Da |
-Macromolecule #7: PYROPHOSPHATE 2-
| Macromolecule | Name: PYROPHOSPHATE 2- / type: ligand / ID: 7 / Number of copies: 1 / Formula: POP |
|---|---|
| Molecular weight | Theoretical: 175.959 Da |
| Chemical component information |  ChemComp-POP: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2 mg/mL | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| |||||||||||||||
| Vitrification | Cryogen name: ETHANE / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 54.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller






















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





















