+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-25780 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of archazolid A bound to yeast VO V-ATPase | |||||||||
 Map data Map data | Yeast VO motor with Archazolid A | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | inhibitor / complex / proton pump / ATPase / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationcell wall mannoprotein biosynthetic process / ATPase-coupled ion transmembrane transporter activity / cellular response to alkaline pH / protein localization to vacuolar membrane / Insulin receptor recycling / Transferrin endocytosis and recycling / polyphosphate metabolic process / ROS and RNS production in phagocytes / Amino acids regulate mTORC1 / Golgi lumen acidification ...cell wall mannoprotein biosynthetic process / ATPase-coupled ion transmembrane transporter activity / cellular response to alkaline pH / protein localization to vacuolar membrane / Insulin receptor recycling / Transferrin endocytosis and recycling / polyphosphate metabolic process / ROS and RNS production in phagocytes / Amino acids regulate mTORC1 / Golgi lumen acidification / P-type proton-exporting transporter activity / vacuolar transport / vacuolar proton-transporting V-type ATPase, V0 domain / endosomal lumen acidification / vacuole organization / protein targeting to vacuole / proton-transporting V-type ATPase complex / fungal-type vacuole / vacuolar proton-transporting V-type ATPase complex / cellular hyperosmotic response / vacuolar acidification / fungal-type vacuole membrane / phosphatidylinositol-3,5-bisphosphate binding / proton transmembrane transporter activity / proton-transporting ATPase activity, rotational mechanism / intracellular copper ion homeostasis / Neutrophil degranulation / RNA endonuclease activity / proton transmembrane transport / cell periphery / transmembrane transport / endocytosis / ATPase binding / protein-containing complex assembly / intracellular iron ion homeostasis / membrane raft / Golgi membrane / endoplasmic reticulum membrane / membrane Similarity search - Function | |||||||||
| Biological species |  | |||||||||
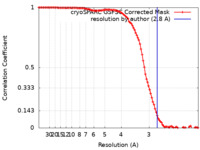
| Method | single particle reconstruction / cryo EM / Resolution: 2.8 Å | |||||||||
 Authors Authors | Keon KA / Rubinstein JL | |||||||||
| Funding support |  Canada, 1 items Canada, 1 items
| |||||||||
 Citation Citation |  Journal: ACS Chem Biol / Year: 2022 Journal: ACS Chem Biol / Year: 2022Title: Cryo-EM of the Yeast V Complex Reveals Distinct Binding Sites for Macrolide V-ATPase Inhibitors. Authors: Kristine A Keon / Samir Benlekbir / Susanne H Kirsch / Rolf Müller / John L Rubinstein /   Abstract: Vacuolar-type adenosine triphosphatases (V-ATPases) are proton pumps found in almost all eukaryotic cells. These enzymes consist of a soluble catalytic V region that hydrolyzes ATP and a membrane- ...Vacuolar-type adenosine triphosphatases (V-ATPases) are proton pumps found in almost all eukaryotic cells. These enzymes consist of a soluble catalytic V region that hydrolyzes ATP and a membrane-embedded V region responsible for proton translocation. V-ATPase activity leads to acidification of endosomes, phagosomes, lysosomes, secretory vesicles, and the trans-Golgi network, with extracellular acidification occurring in some specialized cells. Small-molecule inhibitors of V-ATPase have played a crucial role in elucidating numerous aspects of cell biology by blocking acidification of intracellular compartments, while therapeutic use of V-ATPase inhibitors has been proposed for the treatment of cancer, osteoporosis, and some infections. Here, we determine structures of the isolated V complex from bound to two well-known macrolide inhibitors: bafilomycin A1 and archazolid A. The structures reveal different binding sites for the inhibitors on the surface of the proton-carrying c ring, with only a small amount of overlap between the two sites. Binding of both inhibitors is mediated primarily through van der Waals interactions in shallow pockets and suggests that the inhibitors block rotation of the ring. Together, these structures indicate the existence of a large chemical space available for V-ATPase inhibitors that block acidification by binding the c ring. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_25780.map.gz emd_25780.map.gz | 97.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-25780-v30.xml emd-25780-v30.xml emd-25780.xml emd-25780.xml | 20.5 KB 20.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_25780_fsc.xml emd_25780_fsc.xml | 11.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_25780.png emd_25780.png | 79.3 KB | ||
| Filedesc metadata |  emd-25780.cif.gz emd-25780.cif.gz | 7.3 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-25780 http://ftp.pdbj.org/pub/emdb/structures/EMD-25780 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-25780 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-25780 | HTTPS FTP |
-Related structure data
| Related structure data |  7tapMC  7taoC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_25780.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_25780.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Yeast VO motor with Archazolid A | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.03 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
+Entire : Cryo-EM structure of archazolid A bound to yeast VO V-ATPase
+Supramolecule #1: Cryo-EM structure of archazolid A bound to yeast VO V-ATPase
+Macromolecule #1: V-type proton ATPase subunit c'
+Macromolecule #2: V-type proton ATPase subunit c''
+Macromolecule #3: V0 assembly protein 1
+Macromolecule #4: V-type proton ATPase subunit e
+Macromolecule #5: V-type proton ATPase subunit c
+Macromolecule #6: Yeast V-ATPase subunit f
+Macromolecule #7: V-type proton ATPase subunit d
+Macromolecule #8: V-type proton ATPase subunit a, vacuolar isoform
+Macromolecule #9: Archazolid A
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Average electron dose: 42.0 e/Å2 / Details: Prototype Falcon 4i camera |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5500000000000003 µm / Nominal defocus min: 0.37 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller
























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)