[English] 日本語
 Yorodumi
Yorodumi- EMDB-25709: Rabbit RyR1 disease mutant Y523S in complex with FKBP12.6 embedde... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Rabbit RyR1 disease mutant Y523S in complex with FKBP12.6 embedded in lipidic nanodisc in the closed state | |||||||||||||||
 Map data Map data | ||||||||||||||||
 Sample Sample |
| |||||||||||||||
 Keywords Keywords | Ryanodine receptor / RyR / RyR1 / calcium channel / mutation / NTDC mutation / malignant hyperthermia / central core disease / membrane protein / excitation-contraction coupling | |||||||||||||||
| Function / homology |  Function and homology information Function and homology informationATP-gated ion channel activity / positive regulation of sequestering of calcium ion / negative regulation of calcium-mediated signaling / negative regulation of insulin secretion involved in cellular response to glucose stimulus / neuronal action potential propagation / negative regulation of release of sequestered calcium ion into cytosol / insulin secretion involved in cellular response to glucose stimulus / terminal cisterna / ryanodine-sensitive calcium-release channel activity / ryanodine receptor complex ...ATP-gated ion channel activity / positive regulation of sequestering of calcium ion / negative regulation of calcium-mediated signaling / negative regulation of insulin secretion involved in cellular response to glucose stimulus / neuronal action potential propagation / negative regulation of release of sequestered calcium ion into cytosol / insulin secretion involved in cellular response to glucose stimulus / terminal cisterna / ryanodine-sensitive calcium-release channel activity / ryanodine receptor complex / release of sequestered calcium ion into cytosol by sarcoplasmic reticulum / response to redox state / ossification involved in bone maturation / negative regulation of heart rate / cellular response to caffeine / 'de novo' protein folding / skin development / FK506 binding / organelle membrane / intracellularly gated calcium channel activity / smooth endoplasmic reticulum / outflow tract morphogenesis / smooth muscle contraction / toxic substance binding / T cell proliferation / striated muscle contraction / regulation of cardiac muscle contraction by regulation of the release of sequestered calcium ion / voltage-gated calcium channel activity / calcium channel inhibitor activity / skeletal muscle fiber development / regulation of release of sequestered calcium ion into cytosol by sarcoplasmic reticulum / Ion homeostasis / release of sequestered calcium ion into cytosol / calcium channel complex / sarcoplasmic reticulum membrane / cellular response to calcium ion / muscle contraction / sarcoplasmic reticulum / protein maturation / calcium channel regulator activity / peptidylprolyl isomerase / peptidyl-prolyl cis-trans isomerase activity / calcium-mediated signaling / sarcolemma / calcium ion transmembrane transport / Stimuli-sensing channels / calcium channel activity / Z disc / intracellular calcium ion homeostasis / disordered domain specific binding / positive regulation of cytosolic calcium ion concentration / protein refolding / protein homotetramerization / transmembrane transporter binding / calmodulin binding / signaling receptor binding / calcium ion binding / ATP binding / identical protein binding / membrane / cytoplasm Similarity search - Function | |||||||||||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | |||||||||||||||
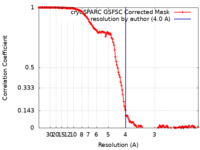
| Method | single particle reconstruction / cryo EM / Resolution: 4.0 Å | |||||||||||||||
 Authors Authors | Iyer KA / Hu Y / Murayama T / Samso M | |||||||||||||||
| Funding support |  United States, 4 items United States, 4 items
| |||||||||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2022 Journal: Proc Natl Acad Sci U S A / Year: 2022Title: Molecular mechanism of the severe MH/CCD mutation Y522S in skeletal ryanodine receptor (RyR1) by cryo-EM. Authors: Kavita A Iyer / Yifan Hu / Thomas Klose / Takashi Murayama / Montserrat Samsó /   Abstract: Ryanodine receptors (RyRs) are main regulators of intracellular Ca release and muscle contraction. The Y522S mutation of RyR1 causes central core disease, a weakening myopathy, and malignant ...Ryanodine receptors (RyRs) are main regulators of intracellular Ca release and muscle contraction. The Y522S mutation of RyR1 causes central core disease, a weakening myopathy, and malignant hyperthermia, a sudden and potentially fatal response to anesthetics or heat. Y522 is in the core of the N-terminal subdomain C of RyR1 and the mechanism of how this mutation orchestrates malfunction is unpredictable for this 2-MDa ion channel, which has four identical subunits composed of 15 distinct cytoplasmic domains each. We expressed and purified the RyR1 rabbit homolog, Y523S, from HEK293 cells and reconstituted it in nanodiscs under closed and open states. The high-resolution cryogenic electron microscopic (cryo-EM) three-dimensional (3D) structures show that the phenyl ring of Tyr functions in a manner analogous to a "spacer" within an α-helical bundle. Mutation to the much smaller Ser alters the hydrophobic network within the bundle, triggering rearrangement of its α-helices with repercussions in the orientation of most cytoplasmic domains. Examining the mutation-induced readjustments exposed a series of connected α-helices acting as an ∼100 Å-long lever: One end protrudes toward the dihydropyridine receptor, its molecular activator (akin to an antenna), while the other end reaches the Ca activation site. The Y523S mutation elicits channel preactivation in the absence of any activator and full opening at 1.5 µM free Ca, increasing by ∼20-fold the potency of Ca to activate the channel compared with RyR1 wild type (WT). This study identified a preactivated pathological state of RyR1 and a long-range lever that may work as a molecular switch to open the channel. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_25709.map.gz emd_25709.map.gz | 286.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-25709-v30.xml emd-25709-v30.xml emd-25709.xml emd-25709.xml | 29.2 KB 29.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_25709_fsc.xml emd_25709_fsc.xml | 14.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_25709.png emd_25709.png | 100.6 KB | ||
| Filedesc metadata |  emd-25709.cif.gz emd-25709.cif.gz | 9.7 KB | ||
| Others |  emd_25709_additional_1.map.gz emd_25709_additional_1.map.gz emd_25709_additional_2.map.gz emd_25709_additional_2.map.gz emd_25709_half_map_1.map.gz emd_25709_half_map_1.map.gz emd_25709_half_map_2.map.gz emd_25709_half_map_2.map.gz | 139.8 MB 286.7 MB 281.3 MB 281.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-25709 http://ftp.pdbj.org/pub/emdb/structures/EMD-25709 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-25709 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-25709 | HTTPS FTP |
-Related structure data
| Related structure data |  7t64MC  7t65C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_25709.map.gz / Format: CCP4 / Size: 303.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_25709.map.gz / Format: CCP4 / Size: 303.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.08 Å | ||||||||||||||||||||||||||||||||||||
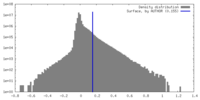
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: Focused map of single subunit generated using local...
| File | emd_25709_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Focused map of single subunit generated using local refinement in cryoSPARC0 | ||||||||||||
| Projections & Slices |
| ||||||||||||
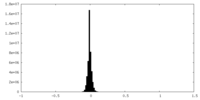
| Density Histograms |
-Additional map: Focused map of NTDs (A, B and C)...
| File | emd_25709_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Focused map of NTDs (A, B and C) and SPRYs (1 to 3) of a single subunit generated using particle subtraction and local refinement in cryoSPARC | ||||||||||||
| Projections & Slices |
| ||||||||||||
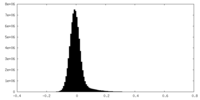
| Density Histograms |
-Half map: #2
| File | emd_25709_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
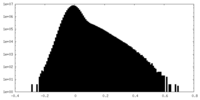
| Density Histograms |
-Half map: #1
| File | emd_25709_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Rabbit RyR1 disease mutant Y523S in complex with FKBP12.6 embedde...
| Entire | Name: Rabbit RyR1 disease mutant Y523S in complex with FKBP12.6 embedded in lipidic nanodisc in the closed state |
|---|---|
| Components |
|
-Supramolecule #1: Rabbit RyR1 disease mutant Y523S in complex with FKBP12.6 embedde...
| Supramolecule | Name: Rabbit RyR1 disease mutant Y523S in complex with FKBP12.6 embedded in lipidic nanodisc in the closed state type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Molecular weight | Theoretical: 2.26 MDa |
-Macromolecule #1: Ryanodine receptor 1
| Macromolecule | Name: Ryanodine receptor 1 / type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 565.832562 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MGDGGEGEDE VQFLRTDDEV VLQCSATVLK EQLKLCLAAE GFGNRLCFLE PTSNAQNVPP DLAICCFTLE QSLSVRALQE MLANTVEAG VESSQGGGHR TLLYGHAILL RHAHSRMYLS CLTTSRSMTD KLAFDVGLQE DATGEACWWT MHPASKQRSE G EKVRVGDD ...String: MGDGGEGEDE VQFLRTDDEV VLQCSATVLK EQLKLCLAAE GFGNRLCFLE PTSNAQNVPP DLAICCFTLE QSLSVRALQE MLANTVEAG VESSQGGGHR TLLYGHAILL RHAHSRMYLS CLTTSRSMTD KLAFDVGLQE DATGEACWWT MHPASKQRSE G EKVRVGDD LILVSVSSER YLHLSTASGE LQVDASFMQT LWNMNPICSC CEEGYVTGGH VLRLFHGHMD ECLTISAADS DD QRRLVYY EGGAVCTHAR SLWRLEPLRI SWSGSHLRWG QPLRIRHVTT GRYLALTEDQ GLVVVDACKA HTKATSFCFR VSK EKLDTA PKRDVEGMGP PEIKYGESLC FVQHVASGLW LTYAAPDPKA LRLGVLKKKA ILHQEGHMDD ALFLTRCQQE ESQA ARMIH STAGLYNQFI KGLDSFSGKP RGSGPPAGPA LPIEAVILSL QDLIGYFEPP SEELQHEEKQ SKLRSLRNRQ SLFQE EGML SLVLNCIDRL NVYTTAAHFA EYAGEEAAES WKEIVNLLSE LLASLIRGNR ANCALFSTNL DWVVSKLDRL EASSGI LEV LYCVLIESPE VLNIIQENHI KSIISLLDKH GRNHKVLDVL CSLCVCNGVA VRSNQDLITE NLLPGRELLL QTNLINY VT SIRPNIFVGR AEGSTQYGKW YFEVMVDEVV PFLTAQATHL RVGWALTEGY SPYPGGGEGW GGNGVGDDLY SYGFDGLH L WTGHVARPVT SPGQHLLAPE DVVSCCLDLS VPSISFRING CPVQGVFEAF NLDGLFFPVV SFSAGVKVRF LLGGRHGEF KFLPPPGYAP CHEAVLPRER LRLEPIKEYR REGPRGPHLV GPSRCLSHTD FVPCPVDTVQ IVLPPHLERI REKLAENIHE LWALTRIEQ GWTYGPVRDD NKRLHPCLVN FHSLPEPERN YNLQMSGETL KTLLALGCHV GMADEKAEDN LKKTKLPKTY M MSNGYKPA PLDLSHVRLT PAQTTLVDRL AENGHNVWAR DRVAQGWSYS AVQDIPARRN PRLVPYRLLD EATKRSNRDS LC QAVRTLL GYGYNIEPPD QEPSQVENQS RWDRVRIFRA EKSYTVQSGR WYFEFEAVTT GEMRVGWARP ELRPDVELGA DEL AYVFNG HRGQRWHLGS EPFGRPWQSG DVVGCMIDLT ENTIIFTLNG EVLMSDSGSE TAFREIEIGD GFLPVCSLGP GQVG HLNLG QDVSSLRFFA ICGLQEGFEP FAINMQRPVT TWFSKSLPQF EPVPPEHPHY EVARMDGTVD TPPCLRLAHR TWGSQ NSLV EMLFLRLSLP VQFHQHFRCT AGATPLAPPG LQPPAEDEAR AAEPDPDYEN LRRSAGGWGE AEGGKEGTAK EGTPGG TPQ PGVEAQPVRA ENEKDATTEK NKKRGFLFKA KKAAMMTQPP ATPALPRLPH DVVPADNRDD PEIILNTTTY YYSVRVF AG QEPSCVWVGW VTPDYHQHDM NFDLSKVRAV TVTMGDEQGN VHSSLKCSNC YMVWGGDFVS PGQQGRISHT DLVIGCLV D LATGLMTFTA NGKESNTFFQ VEPNTKLFPA VFVLPTHQNV IQFELGKQKN IMPLSAAMFL SERKNPAPQC PPRLEVQML MPVSWSRMPN HFLQVETRRA GERLGWAVQC QDPLTMMALH IPEENRCMDI LELSERLDLQ RFHSHTLRLY RAVCALGNNR VAHALCSHV DQAQLLHALE DAHLPGPLRA GYYDLLISIH LESACRSRRS MLSEYIVPLT PETRAITLFP PGRKGGNARR H GLPGVGVT TSLRPPHHFS PPCFVAALPA AGVAEAPARL SPAIPLEALR DKALRMLGEA VRDGGQHARD PVGGSVEFQF VP VLKLVST LLVMGIFGDE DVKQILKMIE PEVFTEEEEE EEEEEEEEEE EEEDEEEKEE DEEEEEKEDA EKEEEEAPEG EKE DLEEGL LQMKLPESVK LQMCNLLEYF CDQELQHRVE SLAAFAERYV DKLQANQRSR YALLMRAFTM SAAETARRTR EFRS PPQEQ INMLLHFKDE ADEEDCPLPE DIRQDLQDFH QDLLAHCGIQ LEGEEEEPEE ETSLSSRLRS LLETVRLVKK KEEKP EEEL PAEEKKPQSL QELVSHMVVR WAQEDYVQSP ELVRAMFSLL HRQYDGLGEL LRALPRAYTI SPSSVEDTMS LLECLG QIR SLLIVQMGPQ EENLMIQSIG NIMNNKVFYQ HPNLMRALGM HETVMEVMVN VLGGGETKEI RFPKMVTSCC RFLCYFC RI SRQNQRSMFD HLSYLLENSG IGLGMQGSTP LDVAAASVID NNELALALQE QDLEKVVSYL AGCGLQSCPM LLAKGYPD I GWNPCGGERY LDFLRFAVFV NGESVEENAN VVVRLLIRKP ECFGPALRGE GGSGLLAAIE EAIRISEDPA RDGPGVRRD RRREHFGEEP PEENRVHLGH AIMSFYAALI DLLGRCAPEM HLIQAGKGEA LRIRAILRSL VPLDDLVGII SLPLQIPTLG KDGALVQPK MSASFVPDHK ASMVLFLDRV YGIENQDFLL HVLDVGFLPD MRAAASLDTA TFSTTEMALA LNRYLCLAVL P LITKCAPL FAGTEHRAIM VDSMLHTVYR LSRGRSLTKA QRDVIEDCLM ALCRYIRPSM LQHLLRRLVF DVPILNEFAK MP LKLLTNH YERCWKYYCL PTGWANFGVT SEEELHLTRK LFWGIFDSLA HKKYDQELYR MAMPCLCAIA GALPPDYVDA SYS SKAEKK ATVDAEGNFD PRPVETLNVI IPEKLDSFIN KFAEYTHEKW AFDKIQNNWS YGENVDEELK THPMLRPYKT FSEK DKEIY RWPIKESLKA MIAWEWTIEK AREGEEERTE KKKTRKISQT AQTYDPREGY NPQPPDLSGV TLSRELQAMA EQLAE NYHN TWGRKKKQEL EAKGGGTHPL LVPYDTLTAK EKARDREKAQ ELLKFLQMNG YAVTRGLKDM ELDTSSIEKR FAFGFL QQL LRWMDISQEF IAHLEAVVSS GRVEKSPHEQ EIKFFAKILL PLINQYFTNH CLYFLSTPAK VLGSGGHASN KEKEMIT SL FCKLAALVRH RVSLFGTDAP AVVNCLHILA RSLDARTVMK SGPEIVKAGL RSFFESASED IEKMVENLRL GKVSQART Q VKGVGQNLTY TTVALLPVLT TLFQHIAQHQ FGDDVILDDV QVSCYRTLCS IYSLGTTKNT YVEKLRPALG ECLARLAAA MPVAFLEPQL NEYNACSVYT TKSPRERAIL GLPNSVEEMC PDIPVLDRLM ADIGGLAESG ARYTEMPHVI EITLPMLCSY LPRWWERGP EAPPPALPAG APPPCTAVTS DHLNSLLGNI LRIIVNNLGI DEATWMKRLA VFAQPIVSRA RPELLHSHFI P TIGRLRKR AGKVVAEEEQ LRLEAKAEAE EGELLVRDEF SVLCRDLYAL YPLLIRYVDN NRAHWLTEPN ANAEELFRMV GE IFIYWSK SHNFKREEQN FVVQNEINNM SFLTADSKSK MAKAGDAQSG GSDQERTKKK RRGDRYSVQT SLIVATLKKM LPI GLNMCA PTDQDLIMLA KTRYALKDTD EEVREFLQNN LHLQGKVEGS PSLRWQMALY RGLPGREEDA DDPEKIVRRV QEVS AVLYH LEQTEHPYKS KKAVWHKLLS KQRRRAVVAC FRMTPLYNLP THRACNMFLE SYKAAWILTE DHSFEDRMID DLSKA GEQE EEEEEVEEKK PDPLHQLVLH FSRTALTEKS KLDEDYLYMA YADIMAKSCH LEEGGENGEA EEEEVEVSFE EKEMEK QRL LYQQSRLHTR GAAEMVLQMI SACKGETGAM VSSTLKLGIS ILNGGNAEVQ QKMLDYLKDK KEVGFFQSIQ ALMQTCS VL DLNAFERQNK AEGLGMVNED GTVINRQNGE KVMADDEFTQ DLFRFLQLLC EGHNNDFQNY LRTQTGNTTT INIIICTV D YLLRLQESIS DFYWYYSGKD VIEEQGKRNF SKAMSVAKQV FNSLTEYIQG PCTGNQQSLA HSRLWDAVVG FLHVFAHMM MKLAQDSSQI ELLKELLDLQ KDMVVMLLSL LEGNVVNGMI ARQMVDMLVE SSSNVEMILK FFDMFLKLKD IVGSEAFQDY VTDPRGLIS KKDFQKAMDS QKQFTGPEIQ FLLSCSEADE NEMINFEEFA NRFQEPARDI GFNVAVLLTN LSEHVPHDPR L RNFLELAE SILEYFRPYL GRIEIMGASR RIERIYFEIS ETNRAQWEMP QVKESKRQFI FDVVNEGGEA EKMELFVSFC ED TIFEMQI AAQISEPEGE PEADEDEGMG EAAAEGAEEG AAGAEGAAGT VAAGATARLA AAAARALRGL SYRSLRRRVR RLR RLTARE AATALAALLW AVVARAGAAG AGAAAGALRL LWGSLFGGGL VEGAKKVTVT ELLAGMPDPT SDEVHGEQPA GPGG DADGA GEGEGEGDAA EGDGDEEVAG HEAGPGGAEG VVAVADGGPF RPEGAGGLGD MGDTTPAEPP TPEGSPILKR KLGVD GEEE ELVPEPEPEP EPEPEKADEE NGEKEEVPEA PPEPPKKAPP SPPAKKEEAG GAGMEFWGEL EVQRVKFLNY LSRNFY TLR FLALFLAFAI NFILLFYKVS DSPPGEDDME GSAAGDLAGA GSGGGSGWGS GAGEEAEGDE DENMVYYFLE ESTGYME PA LWCLSLLHTL VAFLCIIGYN CLKVPLVIFK REKELARKLE FDGLYITEQP GDDDVKGQWD RLVLNTPSFP SNYWDKFV K RKVLDKHGDI FGRERIAELL GMDLASLEIT AHNERKPDPP PGLLTWLMSI DVKYQIWKFG VIFTDNSFLY LGWYMVMSL LGHYNNFFFA AHLLDIAMGV KTLRTILSSV THNGKQLVMT VGLLAVVVYL YTVVAFNFFR KFYNKSEDED EPDMKCDDMM TCYLFHMYV GVRAGGGIGD EIEDPAGDEY ELYRVVFDIT FFFFVIVILL AIIQGLIIDA FGELRDQQEQ VKEDMETKCF I CGIGSDYF DTTPHGFETH TLEEHNLANY MFFLMYLINK DETEHTGQES YVWKMYQERC WDFFPAGDCF RKQYEDQLS UniProtKB: Ryanodine receptor 1 |
-Macromolecule #2: Peptidyl-prolyl cis-trans isomerase FKBP1B
| Macromolecule | Name: Peptidyl-prolyl cis-trans isomerase FKBP1B / type: protein_or_peptide / ID: 2 / Number of copies: 4 / Enantiomer: LEVO / EC number: peptidylprolyl isomerase |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 11.667305 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: GVEIETISPG DGRTFPKKGQ TCVVHYTGML QNGKKFDSSR DRNKPFKFRI GKQEVIKGFE EGAAQMSLGQ RAKLTCTPDV AYGATGHPG VIPPNATLIF DVELLNLE UniProtKB: Peptidyl-prolyl cis-trans isomerase FKBP1B |
-Macromolecule #3: ZINC ION
| Macromolecule | Name: ZINC ION / type: ligand / ID: 3 / Number of copies: 4 / Formula: ZN |
|---|---|
| Molecular weight | Theoretical: 65.409 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 4.5 mg/mL | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.4 Component:
| |||||||||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Mesh: 300 | |||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 277.15 K / Instrument: FEI VITROBOT MARK IV / Details: Blot force 2; blot for 1.0s before plunging. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Number grids imaged: 2 / Number real images: 13171 / Average exposure time: 3.12 sec. / Average electron dose: 53.69 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.7000000000000001 µm / Nominal magnification: 81000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)