[English] 日本語
 Yorodumi
Yorodumi- EMDB-25679: Structure of dimeric phosphorylated Pediculus humanus (Ph) PINK1 ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-25679 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of dimeric phosphorylated Pediculus humanus (Ph) PINK1 with extended alpha-C helix in chain B | |||||||||
 Map data Map data | Dimer map (locally filtered). Generated from 3D variability analysis and local refinement of the dimer. Initial dimer was generated from local refinement of a dimer from the dodecamer. | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | PINK1 / Kinase / Transferase / Mitophagy / Parkinson's Disease / Ubiquitin / Phosphorylation / Phospho-ubiquitin | |||||||||
| Function / homology |  Function and homology information Function and homology informationautophagy of mitochondrion / positive regulation of mitochondrial fission / regulation of apoptotic process / mitochondrial outer membrane / protein kinase activity / non-specific serine/threonine protein kinase / mitochondrial inner membrane / protein serine/threonine kinase activity / ATP binding / metal ion binding / cytosol Similarity search - Function | |||||||||
| Biological species |  Pediculus humanus corporis (human body louse) Pediculus humanus corporis (human body louse) | |||||||||
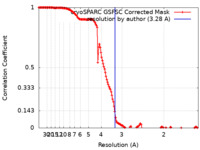
| Method | single particle reconstruction / cryo EM / Resolution: 3.28 Å | |||||||||
 Authors Authors | Gan ZY / Leis A | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Nature / Year: 2022 Journal: Nature / Year: 2022Title: Activation mechanism of PINK1. Authors: Zhong Yan Gan / Sylvie Callegari / Simon A Cobbold / Thomas R Cotton / Michael J Mlodzianoski / Alexander F Schubert / Niall D Geoghegan / Kelly L Rogers / Andrew Leis / Grant Dewson / Alisa ...Authors: Zhong Yan Gan / Sylvie Callegari / Simon A Cobbold / Thomas R Cotton / Michael J Mlodzianoski / Alexander F Schubert / Niall D Geoghegan / Kelly L Rogers / Andrew Leis / Grant Dewson / Alisa Glukhova / David Komander /   Abstract: Mutations in the protein kinase PINK1 lead to defects in mitophagy and cause autosomal recessive early onset Parkinson's disease. PINK1 has many unique features that enable it to phosphorylate ...Mutations in the protein kinase PINK1 lead to defects in mitophagy and cause autosomal recessive early onset Parkinson's disease. PINK1 has many unique features that enable it to phosphorylate ubiquitin and the ubiquitin-like domain of Parkin. Structural analysis of PINK1 from diverse insect species with and without ubiquitin provided snapshots of distinct structural states yet did not explain how PINK1 is activated. Here we elucidate the activation mechanism of PINK1 using crystallography and cryo-electron microscopy (cryo-EM). A crystal structure of unphosphorylated Pediculus humanus corporis (Ph; human body louse) PINK1 resolves an N-terminal helix, revealing the orientation of unphosphorylated yet active PINK1 on the mitochondria. We further provide a cryo-EM structure of a symmetric PhPINK1 dimer trapped during the process of trans-autophosphorylation, as well as a cryo-EM structure of phosphorylated PhPINK1 undergoing a conformational change to an active ubiquitin kinase state. Structures and phosphorylation studies further identify a role for regulatory PINK1 oxidation. Together, our research delineates the complete activation mechanism of PINK1, illuminates how PINK1 interacts with the mitochondrial outer membrane and reveals how PINK1 activity may be modulated by mitochondrial reactive oxygen species. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_25679.map.gz emd_25679.map.gz | 3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-25679-v30.xml emd-25679-v30.xml emd-25679.xml emd-25679.xml | 19.6 KB 19.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_25679_fsc.xml emd_25679_fsc.xml | 16.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_25679.png emd_25679.png | 65.1 KB | ||
| Masks |  emd_25679_msk_1.map emd_25679_msk_1.map | 325 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-25679.cif.gz emd-25679.cif.gz | 6.3 KB | ||
| Others |  emd_25679_additional_1.map.gz emd_25679_additional_1.map.gz emd_25679_half_map_1.map.gz emd_25679_half_map_1.map.gz emd_25679_half_map_2.map.gz emd_25679_half_map_2.map.gz | 161.5 MB 301.1 MB 301.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-25679 http://ftp.pdbj.org/pub/emdb/structures/EMD-25679 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-25679 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-25679 | HTTPS FTP |
-Related structure data
| Related structure data |  7t4lMC  7t3xC  7t4kC  7t4mC  7t4nC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_25679.map.gz / Format: CCP4 / Size: 325 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_25679.map.gz / Format: CCP4 / Size: 325 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Dimer map (locally filtered). Generated from 3D variability analysis and local refinement of the dimer. Initial dimer was generated from local refinement of a dimer from the dodecamer. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.78 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_25679_msk_1.map emd_25679_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Dimer map (unsharpened).
| File | emd_25679_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Dimer map (unsharpened). | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Dimer half map.
| File | emd_25679_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Dimer half map. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Dimer half map.
| File | emd_25679_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Dimer half map. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Dodecameric phosphorylated Pediculus humanus (Ph) PINK1
| Entire | Name: Dodecameric phosphorylated Pediculus humanus (Ph) PINK1 |
|---|---|
| Components |
|
-Supramolecule #1: Dodecameric phosphorylated Pediculus humanus (Ph) PINK1
| Supramolecule | Name: Dodecameric phosphorylated Pediculus humanus (Ph) PINK1 type: complex / ID: 1 / Parent: 0 / Macromolecule list: all Details: Dodecamer was stabilised by hydrogen peroxide treatment prior to phosphorylation and purification by SEC |
|---|---|
| Source (natural) | Organism:  Pediculus humanus corporis (human body louse) Pediculus humanus corporis (human body louse) |
-Macromolecule #1: Serine/threonine-protein kinase PINK1, putative
| Macromolecule | Name: Serine/threonine-protein kinase PINK1, putative / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO / EC number: non-specific serine/threonine protein kinase |
|---|---|
| Source (natural) | Organism:  Pediculus humanus corporis (human body louse) Pediculus humanus corporis (human body louse) |
| Molecular weight | Theoretical: 53.053602 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: GPSGLLTKDD ELEGICWEIR EAVSKGKWND SESENVEQLQ AANLDELDLG EPIAKGCNAV VYSAKLKNVQ SNKLAHQLAV KMMFNYDVE (SEP)NSTAILKAM YRETVPAMSY FFNQNLFNIE NISDFKIRLP PHPNIVRMYS VFADRIPDLQ CNKQLYP EA LPPRINPEGS ...String: GPSGLLTKDD ELEGICWEIR EAVSKGKWND SESENVEQLQ AANLDELDLG EPIAKGCNAV VYSAKLKNVQ SNKLAHQLAV KMMFNYDVE (SEP)NSTAILKAM YRETVPAMSY FFNQNLFNIE NISDFKIRLP PHPNIVRMYS VFADRIPDLQ CNKQLYP EA LPPRINPEGS GRNMSLFLVM KRYDCTLKEY LRDKTPNMRS SILLLSQLLE AVAHMNIHNI SHRDLKSDNI LVDLSEGD A YPTIVITDFG CCLCDKQNGL VIPYRSEDQD KGGNRALMAP EIANAKPGTF SWLNYKKSDL WAVGAIAYEI FNIDNPFYD KTMKLLSKSY KEEDLPELPD TIPFIIRNLV SNMLSRSTNK RLDCDVAATV AQLYLWAPSS WLKENYTLPN SNEIIQWLLC LSSKVLCER DITARNKTNT MSESVSKAQY KGRRSLPEYE LIASFLRRVR LHLVRKGLKW IQELHIYN UniProtKB: Serine/threonine-protein kinase Pink1, mitochondrial |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1.3 mg/mL | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8.5 Component:
| |||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 200 / Support film - Material: CARBON / Support film - topology: HOLEY ARRAY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 60 sec. / Pretreatment - Atmosphere: OTHER / Pretreatment - Pressure: 0.039 kPa | |||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Specialist optics | Energy filter - Slit width: 10 eV |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 1.25 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: OTHER / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.6 µm |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller














 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)