+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-2564 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM of SecA-70S complex | |||||||||
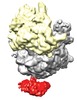
 Map data Map data | reconstruction of SecA-70S complex | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | SecA / ribosome | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 8.8 Å | |||||||||
 Authors Authors | Singh R / Kraft C / Jaiswal R / Sejwal K / Kasaragod V / Kuper J / Buerger J / Mielke T / Luirink J / Bhushan S | |||||||||
 Citation Citation |  Journal: J Biol Chem / Year: 2014 Journal: J Biol Chem / Year: 2014Title: Cryo-electron microscopic structure of SecA protein bound to the 70S ribosome. Authors: Rajkumar Singh / Christian Kraft / Rahul Jaiswal / Kushal Sejwal / Vikram Babu Kasaragod / Jochen Kuper / Jörg Bürger / Thorsten Mielke / Joen Luirink / Shashi Bhushan /    Abstract: SecA is an ATP-dependent molecular motor pumping secretory and outer membrane proteins across the cytoplasmic membrane in bacteria. SecA associates with the protein-conducting channel, the ...SecA is an ATP-dependent molecular motor pumping secretory and outer membrane proteins across the cytoplasmic membrane in bacteria. SecA associates with the protein-conducting channel, the heterotrimeric SecYEG complex, in a so-called posttranslational manner. A recent study further showed binding of a monomeric state of SecA to the ribosome. However, the true oligomeric state of SecA remains controversial because SecA can also form functional dimers, and high-resolution crystal structures exist for both the monomer and the dimer. Here we present the cryo-electron microscopy structures of Escherichia coli SecA bound to the ribosome. We show that not only a monomeric SecA binds to the ribosome but also that two copies of SecA can be observed that form an elongated dimer. Two copies of SecA completely surround the tunnel exit, providing a unique environment to the nascent polypeptides emerging from the ribosome. We identified the N-terminal helix of SecA required for a stable association with the ribosome. The structures indicate a possible function of the dimeric form of SecA at the ribosome. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_2564.map.gz emd_2564.map.gz | 6.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-2564-v30.xml emd-2564-v30.xml emd-2564.xml emd-2564.xml | 9.9 KB 9.9 KB | Display Display |  EMDB header EMDB header |
| Images |  EMD-2564-monomer.tif EMD-2564-monomer.tif | 896.2 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-2564 http://ftp.pdbj.org/pub/emdb/structures/EMD-2564 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-2564 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-2564 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_2564.map.gz / Format: CCP4 / Size: 94.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_2564.map.gz / Format: CCP4 / Size: 94.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | reconstruction of SecA-70S complex | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.24 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : SecA-70S ribosome
| Entire | Name: SecA-70S ribosome |
|---|---|
| Components |
|
-Supramolecule #1000: SecA-70S ribosome
| Supramolecule | Name: SecA-70S ribosome / type: sample / ID: 1000 Oligomeric state: two copies of SecA bound to the 70S ribosome Number unique components: 2 |
|---|---|
| Molecular weight | Experimental: 2 MDa / Theoretical: 2 MDa |
-Supramolecule #1: 70S ribosome
| Supramolecule | Name: 70S ribosome / type: complex / ID: 1 / Name.synonym: 70S / Recombinant expression: No / Ribosome-details: ribosome-prokaryote: ALL |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Experimental: 2 MDa / Theoretical: 2 MDa |
-Macromolecule #1: SecA
| Macromolecule | Name: SecA / type: protein_or_peptide / ID: 1 / Recombinant expression: No |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Experimental: 100 KDa / Theoretical: 100 KDa |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.6 Details: 40 mM Hepes pH 7.6, 50 mM K-acetate, 25 mM Mg-Acetate, 5mM DTT, 0.1% protease inhibitor pill/ml, 0.1 U/ml RNAsin, 125 mM sucrose |
|---|---|
| Grid | Details: 2-nm carbon-coated holey grids |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Instrument: FEI VITROBOT MARK III / Method: Blot for 5 seconds before plunging |
- Electron microscopy
Electron microscopy
| Microscope | FEI POLARA 300 |
|---|---|
| Date | Jun 11, 2011 |
| Image recording | Category: FILM / Film or detector model: KODAK 4489 FILM / Digitization - Scanner: OTHER / Digitization - Sampling interval: 0.25 µm / Number real images: 300 / Average electron dose: 25 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: BRIGHT FIELD / Nominal defocus max: 4.5 µm / Nominal defocus min: 1.5 µm / Nominal magnification: 39000 |
| Sample stage | Specimen holder model: GATAN LIQUID NITROGEN |
| Experimental equipment |  Model: Tecnai Polara / Image courtesy: FEI Company |
- Image processing
Image processing
| Details | Spider |
|---|---|
| CTF correction | Details: Each Micrographs |
| Final reconstruction | Applied symmetry - Point group: C1 (asymmetric) / Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 8.8 Å / Resolution method: OTHER / Software - Name: Spider / Number images used: 115000 |
| Final angle assignment | Details: Spider |
-Atomic model buiding 1
| Initial model | PDB ID: |
|---|---|
| Software | Name:  Chimera Chimera |
| Details | The domains were separately fitted by manual docking using coot and chimera |
| Refinement | Space: REAL / Protocol: RIGID BODY FIT |
 Movie
Movie Controller
Controller














 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)






















