[English] 日本語
 Yorodumi
Yorodumi- EMDB-25550: Map of wt IRES eIF2-containing 48S initiation complex with IRES d... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Map of wt IRES eIF2-containing 48S initiation complex with IRES domain II in position 6. Structure 21(wt) | |||||||||||||||
 Map data Map data | ||||||||||||||||
 Sample Sample |
| |||||||||||||||
 Keywords Keywords | HCV / IRES / 40S / RIBOSOME | |||||||||||||||
| Biological species |  | |||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.76 Å | |||||||||||||||
 Authors Authors | Brown ZP / Abaeva IS / De S / Hellen CUT / Pestova TV / Frank J | |||||||||||||||
| Funding support |  United States, 4 items United States, 4 items
| |||||||||||||||
 Citation Citation |  Journal: EMBO J / Year: 2022 Journal: EMBO J / Year: 2022Title: Molecular architecture of 40S translation initiation complexes on the hepatitis C virus IRES. Authors: Zuben P Brown / Irina S Abaeva / Swastik De / Christopher U T Hellen / Tatyana V Pestova / Joachim Frank /  Abstract: Hepatitis C virus mRNA contains an internal ribosome entry site (IRES) that mediates end-independent translation initiation, requiring a subset of eukaryotic initiation factors (eIFs). Biochemical ...Hepatitis C virus mRNA contains an internal ribosome entry site (IRES) that mediates end-independent translation initiation, requiring a subset of eukaryotic initiation factors (eIFs). Biochemical studies revealed that direct binding of the IRES to the 40S ribosomal subunit places the initiation codon into the P site, where it base pairs with eIF2-bound Met-tRNAiMet forming a 48S initiation complex. Subsequently, eIF5 and eIF5B mediate subunit joining, yielding an elongation-competent 80S ribosome. Initiation can also proceed without eIF2, in which case Met-tRNAiMet is recruited directly by eIF5B. However, the structures of initiation complexes assembled on the HCV IRES, the transitions between different states, and the accompanying conformational changes have remained unknown. To fill these gaps, we now obtained cryo-EM structures of IRES initiation complexes, at resolutions up to 3.5 Å, that cover all major stages from the initial ribosomal association, through eIF2-containing 48S initiation complexes, to eIF5B-containing complexes immediately prior to subunit joining. These structures provide insights into the dynamic network of 40S/IRES contacts, highlight the role of IRES domain II, and reveal conformational changes that occur during the transition from eIF2- to eIF5B-containing 48S complexes and prepare them for subunit joining. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_25550.map.gz emd_25550.map.gz | 226.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-25550-v30.xml emd-25550-v30.xml emd-25550.xml emd-25550.xml | 17.4 KB 17.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_25550_fsc.xml emd_25550_fsc.xml | 14.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_25550.png emd_25550.png | 33.9 KB | ||
| Masks |  emd_25550_msk_1.map emd_25550_msk_1.map | 244.1 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-25550.cif.gz emd-25550.cif.gz | 4.5 KB | ||
| Others |  emd_25550_additional_1.map.gz emd_25550_additional_1.map.gz emd_25550_half_map_1.map.gz emd_25550_half_map_1.map.gz emd_25550_half_map_2.map.gz emd_25550_half_map_2.map.gz | 193 MB 193.8 MB 193.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-25550 http://ftp.pdbj.org/pub/emdb/structures/EMD-25550 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-25550 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-25550 | HTTPS FTP |
-Related structure data
| Related structure data |  7syiC  7syjC  7sykC  7sylC  7syoC  7sypC  7syqC  7syrC  7sysC  7sytC  7syuC  7syvC  7sywC  7syxC C: citing same article ( |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_25550.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_25550.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.95 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_25550_msk_1.map emd_25550_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
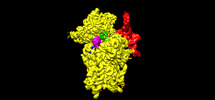
| Projections & Slices |
| ||||||||||||
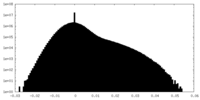
| Density Histograms |
-Additional map: Refine3D map
| File | emd_25550_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Refine3D map | ||||||||||||
| Projections & Slices |
| ||||||||||||
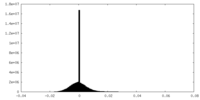
| Density Histograms |
-Half map: #1
| File | emd_25550_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_25550_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : 40S ribosomal small subunit with HCV IRES
| Entire | Name: 40S ribosomal small subunit with HCV IRES |
|---|---|
| Components |
|
-Supramolecule #1: 40S ribosomal small subunit with HCV IRES
| Supramolecule | Name: 40S ribosomal small subunit with HCV IRES / type: complex / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 2 MDa |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.000075 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Grid | Model: Quantifoil R0.6/1 / Material: GOLD / Mesh: 300 / Support film - Material: GOLD / Support film - topology: HOLEY / Support film - Film thickness: 50 / Pretreatment - Type: PLASMA CLEANING / Pretreatment - Time: 25 sec. / Pretreatment - Atmosphere: OTHER / Details: H2/O2 mixture for 25 seconds at 25W power |
| Vitrification | Cryogen name: ETHANE-PROPANE / Chamber humidity: 100 % / Chamber temperature: 277.15 K / Instrument: FEI VITROBOT MARK IV / Details: 4 second blot time, force 3. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI F30 |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average exposure time: 4.0 sec. / Average electron dose: 70.9 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.26 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 52000 |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Tecnai F30 / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller









































 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)