[English] 日本語
 Yorodumi
Yorodumi- EMDB-25389: Structure of the orexin-2 receptor (OX2R) bound to TAK-925, Gi an... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of the orexin-2 receptor (OX2R) bound to TAK-925, Gi and scFv16 | |||||||||
 Map data Map data | Orexin-2 receptor (OX2R) bound to TAK-925, Gi and scFv16 | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | CLASS A GPCR / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationregulation of circadian sleep/wake cycle, wakefulness / circadian sleep/wake cycle process / orexin receptor activity / Orexin and neuropeptides FF and QRFP bind to their respective receptors / Olfactory Signaling Pathway / Sensory perception of sweet, bitter, and umami (glutamate) taste / Synthesis, secretion, and inactivation of Glucagon-like Peptide-1 (GLP-1) / neuropeptide receptor activity / Activation of the phototransduction cascade / feeding behavior ...regulation of circadian sleep/wake cycle, wakefulness / circadian sleep/wake cycle process / orexin receptor activity / Orexin and neuropeptides FF and QRFP bind to their respective receptors / Olfactory Signaling Pathway / Sensory perception of sweet, bitter, and umami (glutamate) taste / Synthesis, secretion, and inactivation of Glucagon-like Peptide-1 (GLP-1) / neuropeptide receptor activity / Activation of the phototransduction cascade / feeding behavior / locomotion / Activation of G protein gated Potassium channels / G-protein activation / G beta:gamma signalling through PI3Kgamma / Prostacyclin signalling through prostacyclin receptor / G beta:gamma signalling through PLC beta / ADP signalling through P2Y purinoceptor 1 / Thromboxane signalling through TP receptor / Presynaptic function of Kainate receptors / G beta:gamma signalling through CDC42 / Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits / G alpha (12/13) signalling events / Glucagon-type ligand receptors / G beta:gamma signalling through BTK / ADP signalling through P2Y purinoceptor 12 / Adrenaline,noradrenaline inhibits insulin secretion / Cooperation of PDCL (PhLP1) and TRiC/CCT in G-protein beta folding / Ca2+ pathway / Thrombin signalling through proteinase activated receptors (PARs) / G alpha (z) signalling events / Extra-nuclear estrogen signaling / G alpha (s) signalling events / G alpha (q) signalling events / G alpha (i) signalling events / Glucagon-like Peptide-1 (GLP1) regulates insulin secretion / High laminar flow shear stress activates signaling by PIEZO1 and PECAM1:CDH5:KDR in endothelial cells / Vasopressin regulates renal water homeostasis via Aquaporins / peptide hormone binding / regulation of cytosolic calcium ion concentration / neuropeptide signaling pathway / cellular response to hormone stimulus / adenylate cyclase inhibitor activity / positive regulation of protein localization to cell cortex / T cell migration / Adenylate cyclase inhibitory pathway / response to prostaglandin E / D2 dopamine receptor binding / G protein-coupled serotonin receptor binding / adenylate cyclase regulator activity / adenylate cyclase-inhibiting serotonin receptor signaling pathway / cellular response to forskolin / regulation of mitotic spindle organization / Regulation of insulin secretion / positive regulation of cholesterol biosynthetic process / negative regulation of insulin secretion / G protein-coupled receptor binding / adenylate cyclase-inhibiting G protein-coupled receptor signaling pathway / response to peptide hormone / adenylate cyclase-modulating G protein-coupled receptor signaling pathway / centriolar satellite / G-protein beta/gamma-subunit complex binding / ADP signalling through P2Y purinoceptor 12 / photoreceptor disc membrane / GDP binding / Adrenaline,noradrenaline inhibits insulin secretion / G alpha (z) signalling events / ADORA2B mediated anti-inflammatory cytokines production / cellular response to catecholamine stimulus / adenylate cyclase-activating dopamine receptor signaling pathway / GPER1 signaling / G-protein beta-subunit binding / cellular response to prostaglandin E stimulus / heterotrimeric G-protein complex / sensory perception of taste / signaling receptor complex adaptor activity / retina development in camera-type eye / G protein activity / GTPase binding / midbody / cell cortex / G alpha (i) signalling events / G alpha (s) signalling events / phospholipase C-activating G protein-coupled receptor signaling pathway / G alpha (q) signalling events / Hydrolases; Acting on acid anhydrides; Acting on GTP to facilitate cellular and subcellular movement / chemical synaptic transmission / Extra-nuclear estrogen signaling / cell population proliferation / ciliary basal body / G protein-coupled receptor signaling pathway / cell division / lysosomal membrane / GTPase activity / synapse / centrosome / GTP binding / protein-containing complex binding / nucleolus / magnesium ion binding / Golgi apparatus Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /  | |||||||||
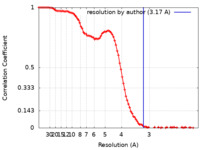
| Method | single particle reconstruction / cryo EM / Resolution: 3.17 Å | |||||||||
 Authors Authors | McGrath AP / Kang Y | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2022 Journal: Nat Commun / Year: 2022Title: Molecular mechanism of the wake-promoting agent TAK-925. Authors: Jie Yin / Yanyong Kang / Aaron P McGrath / Karen Chapman / Megan Sjodt / Eiji Kimura / Atsutoshi Okabe / Tatsuki Koike / Yuhei Miyanohana / Yuji Shimizu / Rameshu Rallabandi / Peng Lian / ...Authors: Jie Yin / Yanyong Kang / Aaron P McGrath / Karen Chapman / Megan Sjodt / Eiji Kimura / Atsutoshi Okabe / Tatsuki Koike / Yuhei Miyanohana / Yuji Shimizu / Rameshu Rallabandi / Peng Lian / Xiaochen Bai / Mack Flinspach / Jef K De Brabander / Daniel M Rosenbaum /    Abstract: The OX orexin receptor (OXR) is a highly expressed G protein-coupled receptor (GPCR) in the brain that regulates wakefulness and circadian rhythms in humans. Antagonism of OXR is a proven therapeutic ...The OX orexin receptor (OXR) is a highly expressed G protein-coupled receptor (GPCR) in the brain that regulates wakefulness and circadian rhythms in humans. Antagonism of OXR is a proven therapeutic strategy for insomnia drugs, and agonism of OXR is a potentially powerful approach for narcolepsy type 1, which is characterized by the death of orexinergic neurons. Until recently, agonism of OXR had been considered 'undruggable.' We harness cryo-electron microscopy of OXR-G protein complexes to determine how the first clinically tested OXR agonist TAK-925 can activate OXR in a highly selective manner. Two structures of TAK-925-bound OXR with either a G mimetic or G reveal that TAK-925 binds at the same site occupied by antagonists, yet interacts with the transmembrane helices to trigger activating microswitches. Our structural and mutagenesis data show that TAK-925's selectivity is mediated by subtle differences between OX and OX receptor subtypes at the orthosteric pocket. Finally, differences in the polarity of interactions at the G protein binding interfaces help to rationalize OXR's coupling selectivity for G signaling. The mechanisms of TAK-925's binding, activation, and selectivity presented herein will aid in understanding the efficacy of small molecule OXR agonists for narcolepsy and other circadian disorders. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_25389.map.gz emd_25389.map.gz | 49.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-25389-v30.xml emd-25389-v30.xml emd-25389.xml emd-25389.xml | 19.6 KB 19.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_25389_fsc.xml emd_25389_fsc.xml | 11.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_25389.png emd_25389.png | 51.4 KB | ||
| Filedesc metadata |  emd-25389.cif.gz emd-25389.cif.gz | 7.2 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-25389 http://ftp.pdbj.org/pub/emdb/structures/EMD-25389 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-25389 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-25389 | HTTPS FTP |
-Related structure data
| Related structure data |  7sqoMC  7sr8C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_25389.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_25389.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Orexin-2 receptor (OX2R) bound to TAK-925, Gi and scFv16 | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.04 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
- Sample components
Sample components
+Entire : Human OX2R in complex with Gai/Gb/Gg-scFv16.
+Supramolecule #1: Human OX2R in complex with Gai/Gb/Gg-scFv16.
+Supramolecule #2: Orexin receptor type 2
+Supramolecule #3: Guanine nucleotide-binding protein G(i) subunit alpha-1
+Supramolecule #4: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1,...
+Supramolecule #5: scFv-16
+Macromolecule #1: Orexin receptor type 2
+Macromolecule #2: Guanine nucleotide-binding protein G(i) subunit alpha-1
+Macromolecule #3: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1
+Macromolecule #4: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2
+Macromolecule #5: scFv-16
+Macromolecule #6: OLEIC ACID
+Macromolecule #7: methyl (2R,3S)-3-[(methanesulfonyl)amino]-2-({[(1s,4S)-4-phenylcy...
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 5 mg/mL |
|---|---|
| Buffer | pH: 7.2 |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Bioquantum |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 43.7 e/Å2 Details: Cryo-EM movie stacks were collected on a Titan Krios microscope operated at 300 kV under EFTEM mode. Nanoprobe with 1um illumination area was used. Data were recorded on a postGIF Gatan K2 ...Details: Cryo-EM movie stacks were collected on a Titan Krios microscope operated at 300 kV under EFTEM mode. Nanoprobe with 1um illumination area was used. Data were recorded on a postGIF Gatan K2 summit camera at a nominal magnification of 130,000, using super-resolution counting model. Bioquantum energy filter was operated in the zero-energy-loss mode with an energy slit width of 20 eV. |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal magnification: 130000 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Protocol: RIGID BODY FIT |
|---|---|
| Output model |  PDB-7sqo: |
 Movie
Movie Controller
Controller























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)




















 Spodoptera (butterflies/moths)
Spodoptera (butterflies/moths)