[English] 日本語
 Yorodumi
Yorodumi- EMDB-2532: Tomographic subvolume average of EFF-1 fusogen on extracellular v... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-2532 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Tomographic subvolume average of EFF-1 fusogen on extracellular vesicles | |||||||||
 Map data Map data | Tomographic subvolume average of EFF-1 on nanotubular extracellular vesicles | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | cell-cell fusion / extracellular fusion / membrane fusion / fusogen / pre-fusion state | |||||||||
| Function / homology |  Function and homology information Function and homology informationnematode male tail mating organ morphogenesis / fusogenic activity / EFF-1 complex / nematode pharyngeal muscle development / post-embryonic body morphogenesis / nematode male tail tip morphogenesis / plasma membrane fusion / cell-cell fusion / vulval development / egg-laying behavior ...nematode male tail mating organ morphogenesis / fusogenic activity / EFF-1 complex / nematode pharyngeal muscle development / post-embryonic body morphogenesis / nematode male tail tip morphogenesis / plasma membrane fusion / cell-cell fusion / vulval development / egg-laying behavior / plasma membrane => GO:0005886 / syncytium formation by plasma membrane fusion / embryonic body morphogenesis / cell-cell contact zone / locomotion / morphogenesis of an epithelium / kinase activity / extracellular region / identical protein binding / plasma membrane / cytoplasm Similarity search - Function | |||||||||
| Biological species |  | |||||||||
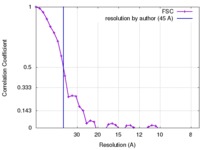
| Method | subtomogram averaging / cryo EM / Resolution: 45.0 Å | |||||||||
 Authors Authors | Zeev-Ben-Mordehai T / Vasishtan D / Siebert CA / Grunewald K | |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2014 Journal: Nat Commun / Year: 2014Title: The full-length cell-cell fusogen EFF-1 is monomeric and upright on the membrane. Authors: Tzviya Zeev-Ben-Mordehai / Daven Vasishtan / C Alistair Siebert / Kay Grünewald /  Abstract: Fusogens are membrane proteins that remodel lipid bilayers to facilitate membrane merging. Although several fusogen ectodomain structures have been solved, structural information on full-length, ...Fusogens are membrane proteins that remodel lipid bilayers to facilitate membrane merging. Although several fusogen ectodomain structures have been solved, structural information on full-length, natively membrane-anchored fusogens is scarce. Here we present the electron cryo microscopy three-dimensional reconstruction of the Caenorhabditis elegans epithelial fusion failure 1 (EFF-1) protein natively anchored in cell-derived membrane vesicles. This reveals a membrane protruding, asymmetric, elongated monomer. Flexible fitting of a protomer of the EFF-1 crystal structure, which is homologous to viral class-II fusion proteins, shows that EFF-1 has a hairpin monomeric conformation before fusion. These structural insights, when combined with our observations of membrane-merging intermediates between vesicles, enable us to propose a model for EFF-1 mediated fusion. This process, involving identical proteins on both membranes to be fused, follows a mechanism that shares features of SNARE-mediated fusion while using the structural building blocks of the unilaterally acting class-II viral fusion proteins. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_2532.map.gz emd_2532.map.gz | 2.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-2532-v30.xml emd-2532-v30.xml emd-2532.xml emd-2532.xml | 10.7 KB 10.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_2532_fsc.xml emd_2532_fsc.xml | 3.9 KB | Display |  FSC data file FSC data file |
| Images |  EMD-2532.png EMD-2532.png | 65.6 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-2532 http://ftp.pdbj.org/pub/emdb/structures/EMD-2532 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-2532 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-2532 | HTTPS FTP |
-Validation report
| Summary document |  emd_2532_validation.pdf.gz emd_2532_validation.pdf.gz | 234.2 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_2532_full_validation.pdf.gz emd_2532_full_validation.pdf.gz | 233.4 KB | Display | |
| Data in XML |  emd_2532_validation.xml.gz emd_2532_validation.xml.gz | 7.3 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-2532 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-2532 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-2532 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-2532 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_2532.map.gz / Format: CCP4 / Size: 2.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_2532.map.gz / Format: CCP4 / Size: 2.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Tomographic subvolume average of EFF-1 on nanotubular extracellular vesicles | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 3.8 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Epithelial fusion failure 1 (EFF-1) Isoform A on extracellular ve...
| Entire | Name: Epithelial fusion failure 1 (EFF-1) Isoform A on extracellular vesicles |
|---|---|
| Components |
|
-Supramolecule #1000: Epithelial fusion failure 1 (EFF-1) Isoform A on extracellular ve...
| Supramolecule | Name: Epithelial fusion failure 1 (EFF-1) Isoform A on extracellular vesicles type: sample / ID: 1000 / Oligomeric state: Monomer / Number unique components: 1 |
|---|---|
| Molecular weight | Theoretical: 87 KDa |
-Macromolecule #1: Epithelial fusion failure 1, isoform a
| Macromolecule | Name: Epithelial fusion failure 1, isoform a / type: protein_or_peptide / ID: 1 / Name.synonym: EFF-1a / Details: Proteins on extracellular vesicles / Number of copies: 1 / Oligomeric state: Monomer / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 87 KDa |
| Recombinant expression | Organism:  |
| Sequence | UniProtKB: EFF-1A GO: plasma membrane fusion, syncytium formation by plasma membrane fusion, plasma membrane => GO:0005886, extracellular region, identical protein binding |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | subtomogram averaging |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 / Details: 25mM HEPES, 130mM NaCl |
|---|---|
| Grid | Details: Holey carbon on top of 200 mesh gold grid. |
| Vitrification | Cryogen name: ETHANE-PROPANE MIXTURE / Chamber temperature: 77 K / Instrument: HOMEMADE PLUNGER / Method: Blot for 3 seconds before plunging |
- Electron microscopy
Electron microscopy
| Microscope | FEI POLARA 300 |
|---|---|
| Temperature | Min: 80 K / Max: 100 K / Average: 85 K |
| Alignment procedure | Legacy - Astigmatism: Objective lens astigmatism was corrected at 115,000 times magnification Legacy - Electron beam tilt params: +1 to -1 mradians |
| Specialist optics | Energy filter - Name: Gatan Quantum 964 / Energy filter - Lower energy threshold: 0.0 eV / Energy filter - Upper energy threshold: 20.0 eV |
| Date | Mar 3, 2013 |
| Image recording | Category: CCD / Film or detector model: GATAN ULTRASCAN 4000 (4k x 4k) / Digitization - Sampling interval: 30 µm / Number real images: 39 / Average electron dose: 60 e/Å2 / Details: 1 tomogram created from 39 projection images / Bits/pixel: 16 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 78950 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 2.0 µm / Nominal magnification: 95000 |
| Sample stage | Specimen holder: Liquid nitrogen cooled / Specimen holder model: GATAN HELIUM / Tilt series - Axis1 - Min angle: -60 ° / Tilt series - Axis1 - Max angle: 60 ° |
| Experimental equipment |  Model: Tecnai Polara / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller
















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)