+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of G6PD-D200N tetramer bound to NADP+ and G6P | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | D200N mutant / OXIDOREDUCTASE | |||||||||
| Function / homology |  Function and homology information Function and homology informationpentose biosynthetic process / ribose phosphate biosynthetic process / response to iron(III) ion / positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel / glucose-6-phosphate dehydrogenase (NADP+) / glucose-6-phosphate dehydrogenase activity / Pentose phosphate pathway / pentose-phosphate shunt, oxidative branch / negative regulation of cell growth involved in cardiac muscle cell development / NADPH regeneration ...pentose biosynthetic process / ribose phosphate biosynthetic process / response to iron(III) ion / positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel / glucose-6-phosphate dehydrogenase (NADP+) / glucose-6-phosphate dehydrogenase activity / Pentose phosphate pathway / pentose-phosphate shunt, oxidative branch / negative regulation of cell growth involved in cardiac muscle cell development / NADPH regeneration / glucose 6-phosphate metabolic process / NADP+ metabolic process / pentose-phosphate shunt / D-glucose binding / NFE2L2 regulates pentose phosphate pathway genes / response to food / erythrocyte maturation / cholesterol biosynthetic process / negative regulation of reactive oxygen species metabolic process / regulation of neuron apoptotic process / glutathione metabolic process / substantia nigra development / TP53 Regulates Metabolic Genes / lipid metabolic process / cytoplasmic side of plasma membrane / centriolar satellite / glucose metabolic process / NADP binding / cellular response to oxidative stress / response to ethanol / intracellular membrane-bounded organelle / protein homodimerization activity / extracellular exosome / identical protein binding / membrane / cytosol / cytoplasm Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.5 Å | |||||||||
 Authors Authors | Wei X / Marmorstein R | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2022 Journal: Proc Natl Acad Sci U S A / Year: 2022Title: Allosteric role of a structural NADP molecule in glucose-6-phosphate dehydrogenase activity. Authors: Xuepeng Wei / Kathryn Kixmoeller / Elana Baltrusaitis / Xiaolu Yang / Ronen Marmorstein /  Abstract: Human glucose-6-phosphate dehydrogenase (G6PD) is the main cellular source of NADPH, and thus plays a key role in maintaining reduced glutathione to protect cells from oxidative stress disorders such ...Human glucose-6-phosphate dehydrogenase (G6PD) is the main cellular source of NADPH, and thus plays a key role in maintaining reduced glutathione to protect cells from oxidative stress disorders such as hemolytic anemia. G6PD is a multimeric enzyme that uses the cofactors β-D-glucose 6-phosphate (G6P) and "catalytic" NADP (NADPc), as well as a "structural" NADP (NADPs) located ∼25 Å from the active site, to generate NADPH. While X-ray crystallographic and biochemical studies have revealed a role for NADPs in maintaining the catalytic activity by stabilizing the multimeric G6PD conformation, other potential roles for NADPs have not been evaluated. Here, we determined the high resolution cryo-electron microscopy structures of human wild-type G6PD in the absence of bound ligands and a catalytic G6PD-D200N mutant bound to NADPc and NADPs in the absence or presence of G6P. A comparison of these structures, together with previously reported structures, reveals that the unliganded human G6PD forms a mixture of dimers and tetramers with similar overall folds, and binding of NADPs induces a structural ordering of a C-terminal extension region and allosterically regulates G6P binding and catalysis. These studies have implications for understanding G6PD deficiencies and for therapy of G6PD-mediated disorders. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_25227.map.gz emd_25227.map.gz | 20.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-25227-v30.xml emd-25227-v30.xml emd-25227.xml emd-25227.xml | 13.3 KB 13.3 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_25227.png emd_25227.png | 160.3 KB | ||
| Filedesc metadata |  emd-25227.cif.gz emd-25227.cif.gz | 6.1 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-25227 http://ftp.pdbj.org/pub/emdb/structures/EMD-25227 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-25227 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-25227 | HTTPS FTP |
-Validation report
| Summary document |  emd_25227_validation.pdf.gz emd_25227_validation.pdf.gz | 600 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_25227_full_validation.pdf.gz emd_25227_full_validation.pdf.gz | 599.6 KB | Display | |
| Data in XML |  emd_25227_validation.xml.gz emd_25227_validation.xml.gz | 5.8 KB | Display | |
| Data in CIF |  emd_25227_validation.cif.gz emd_25227_validation.cif.gz | 6.6 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-25227 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-25227 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-25227 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-25227 | HTTPS FTP |
-Related structure data
| Related structure data |  7sniMC  7snfC  7sngC  7snhC  7toeC  7tofC  7ualC  7uc2C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_25227.map.gz / Format: CCP4 / Size: 22.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_25227.map.gz / Format: CCP4 / Size: 22.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.83 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
- Sample components
Sample components
-Entire : G6PD protein
| Entire | Name: G6PD protein |
|---|---|
| Components |
|
-Supramolecule #1: G6PD protein
| Supramolecule | Name: G6PD protein / type: organelle_or_cellular_component / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 110 KDa |
-Macromolecule #1: Glucose-6-phosphate 1-dehydrogenase
| Macromolecule | Name: Glucose-6-phosphate 1-dehydrogenase / type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO / EC number: glucose-6-phosphate dehydrogenase (NADP+) |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 60.402762 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MAEQVALSRT QVCGILREEL FQGDAFHQSD THIFIIMGAS GDLAKKKIYP TIWWLFRDGL LPENTFIVGY ARSRLTVADI RKQSEPFFK ATPEEKLKLE DFFARNSYVA GQYDDAASYQ RLNSHMNALH LGSQANRLFY LALPPTVYEA VTKNIHESCM S QIGWNRII ...String: MAEQVALSRT QVCGILREEL FQGDAFHQSD THIFIIMGAS GDLAKKKIYP TIWWLFRDGL LPENTFIVGY ARSRLTVADI RKQSEPFFK ATPEEKLKLE DFFARNSYVA GQYDDAASYQ RLNSHMNALH LGSQANRLFY LALPPTVYEA VTKNIHESCM S QIGWNRII VEKPFGRDLQ SSDRLSNHIS SLFREDQIYR INHYLGKEMV QNLMVLRFAN RIFGPIWNRD NIACVILTFK EP FGTEGRG GYFDEFGIIR DVMQNHLLQM LCLVAMEKPA STNSDDVRDE KVKVLKCISE VQANNVVLGQ YVGNPDGEGE ATK GYLDDP TVPRGSTTAT FAAVVLYVEN ERWDGVPFIL RCGKALNERK AEVRLQFHDV AGDIFHQQCK RNELVIRVQP NEAV YTKMM TKKPGMFFNP EESELDLTYG NRYKNVKLPD AYERLILDVF CGSQMHFVRS DELREAWRIF TPLLHQIELE KPKPI PYIY GSRGPTEADE LMKRVGFQYE GTYKWVNPHK LLEHHHHHH UniProtKB: Glucose-6-phosphate 1-dehydrogenase |
-Macromolecule #2: NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE
| Macromolecule | Name: NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE / type: ligand / ID: 2 / Number of copies: 8 / Formula: NAP |
|---|---|
| Molecular weight | Theoretical: 743.405 Da |
| Chemical component information |  ChemComp-NAP: |
-Macromolecule #3: 6-O-phosphono-beta-D-glucopyranose
| Macromolecule | Name: 6-O-phosphono-beta-D-glucopyranose / type: ligand / ID: 3 / Number of copies: 4 / Formula: BG6 |
|---|---|
| Molecular weight | Theoretical: 260.136 Da |
| Chemical component information | 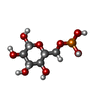 ChemComp-BG6: |
-Macromolecule #4: water
| Macromolecule | Name: water / type: ligand / ID: 4 / Number of copies: 233 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





















