[English] 日本語
 Yorodumi
Yorodumi- EMDB-2522: Substrate Recruitment Pathways in the Yeast Exosome by Electron M... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-2522 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Substrate Recruitment Pathways in the Yeast Exosome by Electron Microscopy | |||||||||
 Map data Map data | Exosome incubated with HDV | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | exosome / RNA degradation route | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / negative staining / Resolution: 23.0 Å | |||||||||
 Authors Authors | Liu J-J / Bratkowski MA / Liu XQ / Niu C-Y / Ke AL / Wang H-W | |||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2014 Journal: Nat Struct Mol Biol / Year: 2014Title: Visualization of distinct substrate-recruitment pathways in the yeast exosome by EM. Authors: Jun-Jie Liu / Matthew A Bratkowski / Xueqi Liu / Chu-Ya Niu / Ailong Ke / Hong-Wei Wang /   Abstract: The eukaryotic exosome is a multisubunit complex typically composed of a catalytically inactive core and the Rrp44 protein, which contains 3'-to-5' exo- and endo-RNase activities. RNA substrates have ...The eukaryotic exosome is a multisubunit complex typically composed of a catalytically inactive core and the Rrp44 protein, which contains 3'-to-5' exo- and endo-RNase activities. RNA substrates have been shown to be recruited through the core to reach Rrp44's exo-RNase (EXO) site. Using single-particle EM and biochemical analysis, we provide visual evidence that two distinct substrate-recruitment pathways exist. In the through-core route, channeling of the single-stranded substrates from the core to Rrp44 induces a characteristic conformational change in Rrp44. In the alternative direct-access route, this conformational change does not take place, and the RNA substrate is visualized to avoid the core and enter Rrp44's EXO site directly. Our results provide mechanistic explanations for several RNA processing scenarios by the eukaryotic exosome and indicate substrate-specific modes of degradation by this complex. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_2522.map.gz emd_2522.map.gz | 948 KB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-2522-v30.xml emd-2522-v30.xml emd-2522.xml emd-2522.xml | 8 KB 8 KB | Display Display |  EMDB header EMDB header |
| Images |  EMD-2522-RE-HDV40.png EMD-2522-RE-HDV40.png | 44.3 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-2522 http://ftp.pdbj.org/pub/emdb/structures/EMD-2522 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-2522 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-2522 | HTTPS FTP |
-Related structure data
| Related structure data |  2491C  2492C  2493C  2494C  2495C  2496C  2497C  2498C 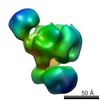 2499C 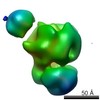 2500C  2501C  2502C C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_2522.map.gz / Format: CCP4 / Size: 1001 KB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_2522.map.gz / Format: CCP4 / Size: 1001 KB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Exosome incubated with HDV | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 4.4 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
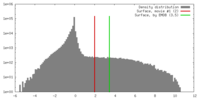
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : HDV-RE
| Entire | Name: HDV-RE |
|---|---|
| Components |
|
-Supramolecule #1000: HDV-RE
| Supramolecule | Name: HDV-RE / type: sample / ID: 1000 / Number unique components: 1 |
|---|---|
| Molecular weight | Experimental: 350 KDa / Method: Mass spectrometry |
-Macromolecule #1: Rrp44
| Macromolecule | Name: Rrp44 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Recombinant expression: No |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Experimental: 110 KDa |
-Experimental details
-Structure determination
| Method | negative staining |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1 mg/mL |
|---|---|
| Buffer | pH: 8 / Details: 150mM NaCl, 50mM Tris-HCL,1mM DTT |
| Staining | Type: NEGATIVE Details: All samples were diluted to a final concentration of ~80 nM of the exosome in the non-digestive buffer and negatively stained in 2% (w/v) uranyl acetate solution following the standard deep ...Details: All samples were diluted to a final concentration of ~80 nM of the exosome in the non-digestive buffer and negatively stained in 2% (w/v) uranyl acetate solution following the standard deep stain procedure on holey-carbon coated EM copper grids covered with a thin layer of continuous carbon |
| Grid | Details: 300 mesh continus carbon |
| Vitrification | Cryogen name: NONE / Instrument: OTHER |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI 12 |
|---|---|
| Date | Oct 10, 2011 |
| Image recording | Category: CCD / Film or detector model: GATAN ULTRASCAN 4000 (4k x 4k) |
| Electron beam | Acceleration voltage: 120 kV / Electron source: LAB6 |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.5 µm / Nominal defocus min: 0.7 µm / Nominal magnification: 52000 |
| Sample stage | Specimen holder model: OTHER |
- Image processing
Image processing
| Final reconstruction | Applied symmetry - Point group: C1 (asymmetric) / Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 23.0 Å / Resolution method: OTHER / Software - Name: Spider / Number images used: 20000 |
|---|---|
| Final angle assignment | Details: SPIDER: theta 90 degrees, phi 359.9 degrees |
 Movie
Movie Controller
Controller


 UCSF Chimera
UCSF Chimera




 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)