[English] 日本語
 Yorodumi
Yorodumi- EMDB-24882: Human Nuclear exosome targeting (NEXT) complex homodimer bound to... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Human Nuclear exosome targeting (NEXT) complex homodimer bound to RNA (substrate 1) | |||||||||
 Map data Map data | human NEXT dimer bound to RNA - composite structure | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Helicase / ATPase / RNA / Exosome / RNA BINDING PROTEIN / RNA BINDING PROTEIN-RNA complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationco-transcriptional lncRNA 3' end processing, cleavage and polyadenylation pathway / snRNA catabolic process / TRAMP complex / snRNA binding / mRNA 3'-end processing / RNA catabolic process / maturation of 5.8S rRNA / regulation of alternative mRNA splicing, via spliceosome / pre-mRNA intronic binding / RNA processing ...co-transcriptional lncRNA 3' end processing, cleavage and polyadenylation pathway / snRNA catabolic process / TRAMP complex / snRNA binding / mRNA 3'-end processing / RNA catabolic process / maturation of 5.8S rRNA / regulation of alternative mRNA splicing, via spliceosome / pre-mRNA intronic binding / RNA processing / Major pathway of rRNA processing in the nucleolus and cytosol / 14-3-3 protein binding / catalytic step 2 spliceosome / mRNA Splicing - Major Pathway / Regulation of endogenous retroelements by the Human Silencing Hub (HUSH) complex / meiotic cell cycle / mRNA splicing, via spliceosome / rRNA processing / RNA helicase activity / single-stranded RNA binding / nuclear speck / nuclear body / RNA helicase / DNA damage response / nucleolus / ATP hydrolysis activity / RNA binding / zinc ion binding / nucleoplasm / ATP binding / nucleus Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) / synthetic construct (others) Homo sapiens (human) / synthetic construct (others) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 4.06 Å | |||||||||
 Authors Authors | Puno MR / Lima CD | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Cell / Year: 2022 Journal: Cell / Year: 2022Title: Structural basis for RNA surveillance by the human nuclear exosome targeting (NEXT) complex. Authors: M Rhyan Puno / Christopher D Lima /  Abstract: RNA quality control relies on co-factors and adaptors to identify and prepare substrates for degradation by ribonucleases such as the 3' to 5' ribonucleolytic RNA exosome. Here, we determined ...RNA quality control relies on co-factors and adaptors to identify and prepare substrates for degradation by ribonucleases such as the 3' to 5' ribonucleolytic RNA exosome. Here, we determined cryogenic electron microscopy structures of human nuclear exosome targeting (NEXT) complexes bound to RNA that reveal mechanistic insights to substrate recognition and early steps that precede RNA handover to the exosome. The structures illuminate ZCCHC8 as a scaffold, mediating homodimerization while embracing the MTR4 helicase and flexibly anchoring RBM7 to the helicase core. All three subunits collaborate to bind the RNA, with RBM7 and ZCCHC8 surveying sequences upstream of the 3' end to facilitate RNA capture by MTR4. ZCCHC8 obscures MTR4 surfaces important for RNA binding and extrusion as well as MPP6-dependent recruitment and docking onto the RNA exosome core, interactions that contribute to RNA surveillance by coordinating RNA capture, translocation, and extrusion from the helicase to the exosome for decay. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_24882.map.gz emd_24882.map.gz | 19.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-24882-v30.xml emd-24882-v30.xml emd-24882.xml emd-24882.xml | 53.9 KB 53.9 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_24882.png emd_24882.png | 60.6 KB | ||
| Filedesc metadata |  emd-24882.cif.gz emd-24882.cif.gz | 8.2 KB | ||
| Others |  emd_24882_additional_1.map.gz emd_24882_additional_1.map.gz emd_24882_additional_10.map.gz emd_24882_additional_10.map.gz emd_24882_additional_11.map.gz emd_24882_additional_11.map.gz emd_24882_additional_12.map.gz emd_24882_additional_12.map.gz emd_24882_additional_13.map.gz emd_24882_additional_13.map.gz emd_24882_additional_14.map.gz emd_24882_additional_14.map.gz emd_24882_additional_15.map.gz emd_24882_additional_15.map.gz emd_24882_additional_16.map.gz emd_24882_additional_16.map.gz emd_24882_additional_17.map.gz emd_24882_additional_17.map.gz emd_24882_additional_18.map.gz emd_24882_additional_18.map.gz emd_24882_additional_2.map.gz emd_24882_additional_2.map.gz emd_24882_additional_3.map.gz emd_24882_additional_3.map.gz emd_24882_additional_4.map.gz emd_24882_additional_4.map.gz emd_24882_additional_5.map.gz emd_24882_additional_5.map.gz emd_24882_additional_6.map.gz emd_24882_additional_6.map.gz emd_24882_additional_7.map.gz emd_24882_additional_7.map.gz emd_24882_additional_8.map.gz emd_24882_additional_8.map.gz emd_24882_additional_9.map.gz emd_24882_additional_9.map.gz | 171.1 MB 172 MB 172 MB 171.3 MB 171.7 MB 171.5 MB 171.2 MB 171.4 MB 10.6 MB 171.8 MB 11.4 MB 11.8 MB 11.1 MB 171.7 MB 171.3 MB 171.5 MB 15.6 MB 11.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-24882 http://ftp.pdbj.org/pub/emdb/structures/EMD-24882 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-24882 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-24882 | HTTPS FTP |
-Related structure data
| Related structure data |  7s7bMC  7s7cC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_24882.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_24882.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | human NEXT dimer bound to RNA - composite structure | ||||||||||||||||||||||||||||||||||||
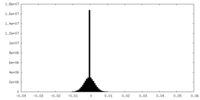
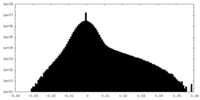
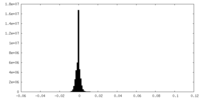
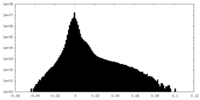
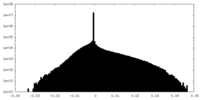
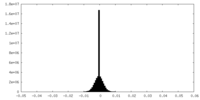
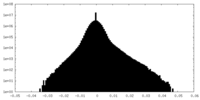
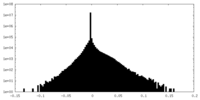
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.088 Å | ||||||||||||||||||||||||||||||||||||
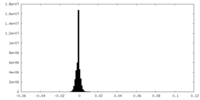
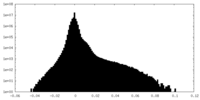
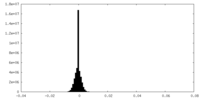
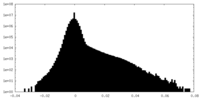
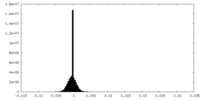
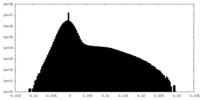
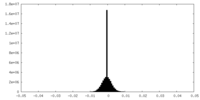
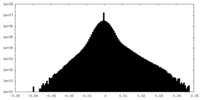
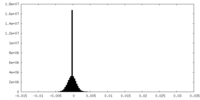
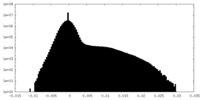
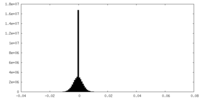
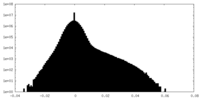
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
+Additional map: focused refinement on protomer A MTR4 core, ZCCHC8...
+Additional map: focused refinement on ZCCHC8-HD/KID MTR4-KOW half map 1
+Additional map: focused refinement on ZCCHC8-HD/KID MTR4-KOW half map 2
+Additional map: focused refinement on protomer A MTR4 half map 1
+Additional map: Overall reconstruction half map 1
+Additional map: focused refinement on protomer B MTR4 core, ZCCHC8...
+Additional map: Overall reconstruction half map 2
+Additional map: focused refinement on protomer A MTR4 core, ZCCHC8...
+Additional map: focused refinement on protomer B MTR4 core, ZCCHC8...
+Additional map: focused refinement on protomer B MTR4 core, ZCCHC8...
+Additional map: focused refinement on protomer B MTR4 (component map...
+Additional map: focused refinement on ZCCHC8-HD/KID MTR4-KOW (component map of...
+Additional map: focused refinement on protomer A MTR4 core, ZCCHC8...
+Additional map: focused refinement on protomer B MTR4 half map 1
+Additional map: focused refinement on protomer A MTR4 half map 2
+Additional map: focused refinement on protomer B MTR4 half map 2
+Additional map: Overall reconstruction (component map of composite structure)
+Additional map: focused refinement on protomer A MTR4 (component map...
- Sample components
Sample components
-Entire : human Nuclear Exosome Targeting (NEXT)-RNA substrate 1 complex
| Entire | Name: human Nuclear Exosome Targeting (NEXT)-RNA substrate 1 complex |
|---|---|
| Components |
|
-Supramolecule #1: human Nuclear Exosome Targeting (NEXT)-RNA substrate 1 complex
| Supramolecule | Name: human Nuclear Exosome Targeting (NEXT)-RNA substrate 1 complex type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#4 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Exosome RNA helicase MTR4
| Macromolecule | Name: Exosome RNA helicase MTR4 / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO / EC number: RNA helicase |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 118.224961 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: SGDMADAFGD ELFSVFEGDS TTAAGTKKDK EKDKGKWKGP PGSADKAGKR FDGKLQSEST NNGKNKRDVD FEGTDEPIFG KKPRIEESI TEDLSLADLM PRVKVQSVET VEGCTHEVAL PAEEDYLPLK PRVGKAAKEY PFILDAFQRE AIQCVDNNQS V LVSAHTSA ...String: SGDMADAFGD ELFSVFEGDS TTAAGTKKDK EKDKGKWKGP PGSADKAGKR FDGKLQSEST NNGKNKRDVD FEGTDEPIFG KKPRIEESI TEDLSLADLM PRVKVQSVET VEGCTHEVAL PAEEDYLPLK PRVGKAAKEY PFILDAFQRE AIQCVDNNQS V LVSAHTSA GKTVCAEYAI ALALREKQRV IFTSPIKALS NQKYREMYEE FQDVGLMTGD VTINPTASCL VMTTEILRSM LY RGSEVMR EVAWVIFDEI HYMRDSERGV VWEETIILLP DNVHYVFLSA TIPNARQFAE WICHLHKQPC HVIYTDYRPT PLQ HYIFPA GGDGLHLVVD ENGDFREDNF NTAMQVLRDA GDLAKGDQKG RKGGTKGPSN VFKIVKMIME RNFQPVIIFS FSKK DCEAY ALQMTKLDFN TDEEKKMVEE VFSNAIDCLS DEDKKLPQVE HVLPLLKRGI GIHHGGLLPI LKETIEILFS EGLIK ALFA TETFAMGINM PARTVLFTNA RKFDGKDFRW ISSGEYIQMS GRAGRRGMDD RGIVILMVDE KMSPTIGKQL LKGSAD PLN SAFHLTYNMV LNLLRVEEIN PEYMLEKSFY QFQHYRAIPG VVEKVKNSEE QYNKIVIPNE ESVVIYYKIR QQLAKLG KE IEEYIHKPKY CLPFLQPGRL VKVKNEGDDF GWGVVVNFSK KSNVKPNSGE LDPLYVVEVL LRCSKESLKN SATEAAKP A KPDEKGEMQV VPVLVHLLSA ISSVRLYIPK DLRPVDNRQS VLKSIQEVQK RFPDGIPLLD PIDDMGIQDQ GLKKVIQKV EAFEHRMYSH PLHNDPNLET VYTLCEKKAQ IAIDIKSAKR ELKKARTVLQ MDELKCRKRV LRRLGFATSS DVIEMKGRVA CEISSADEL LLTEMMFNGL FNDLSAEQAT ALLSCFVFQE NSSEMPKLTE QLAGPLRQMQ ECAKRIAKVS AEAKLEIDEE T YLSSFKPH LMDVVYTWAT GATFAHICKM TDVFEGSIIR CMRRLEELLR QMCQAAKAIG NTELENKFAE GITKIKRDIV FA ASLYL UniProtKB: Exosome RNA helicase MTR4 |
-Macromolecule #2: Zinc finger CCHC domain-containing protein 8,Zinc finger CCHC dom...
| Macromolecule | Name: Zinc finger CCHC domain-containing protein 8,Zinc finger CCHC domain-containing protein 8 type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 69.259805 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: SGDMAAEVYF GDLELFEPFD HPEESIPKPV HTRFKDDDGD EEDENGVGDA ELRERLRQCE ETIEQLRAEN QELKRKLNIL TRPSGILVN DTKLDGPILQ ILFMNNAISK QYHQEIEEFV SNLVKRFEEQ QKNDVEKTSF NLLPQPSSIV LEEDHKVEES C AIKNNKEA ...String: SGDMAAEVYF GDLELFEPFD HPEESIPKPV HTRFKDDDGD EEDENGVGDA ELRERLRQCE ETIEQLRAEN QELKRKLNIL TRPSGILVN DTKLDGPILQ ILFMNNAISK QYHQEIEEFV SNLVKRFEEQ QKNDVEKTSF NLLPQPSSIV LEEDHKVEES C AIKNNKEA FSVVGSVLYF TNFCLDKLGQ PLLNENPQLS EGWEIPKYHQ VFSHIVSLEG QEIQVKAKRP KPHCFNCGSE EH QMKDCPM PRNAARISEK RKEYMDACGE ANNQNFQQRY HAEEVEERFG RFKPGVISEE LQDALGVTDK SLPPFIYRMR QLG YPPGWL KEAELENSGL ALYDGKDGTD GETEVGEIQQ NKSVTYDLSK LVNYPGFNIS TPRGIPDEWR IFGSIPMQAC QQKD VFANY LTSNFQAPGV KSGGAVDEDA LTLEELEEQQ RRIWAALEQA ESVNSDSDVP VDTPLTGNSV ASSPCPNELD LPVPE GKTS EKQTLDEPEV PEIFTKKSEA GHASSPDSEV TSLCQKEKAE LAPVNTEGAL LDNGSVVPNC DISNGGSQKL FPADTS PST ATKIHSPIPD MSKFATGITP FEFENMAEST GMYLRIRSLL KNSPRNQQKN KKASE UniProtKB: Zinc finger CCHC domain-containing protein 8, Zinc finger CCHC domain-containing protein 8 |
-Macromolecule #3: RNA-binding protein 7
| Macromolecule | Name: RNA-binding protein 7 / type: protein_or_peptide / ID: 3 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 9.462986 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: SGDEADRTLF VGNLETKVTE ELLFELFHQA GPVIKVKIPK DKDGKPKQFA FVNFKHEVSV PYAMNLLNGI KLYGRPIKIQ FRS UniProtKB: RNA-binding protein 7 |
-Macromolecule #4: RNA (46-MER)
| Macromolecule | Name: RNA (46-MER) / type: rna / ID: 4 Details: The actual RNA sequence is: ACAUGAGGAUCACCCAUGUAAUCUCUUUCAAAAAA(2PU)ACAAAAAAAA. (2PU) is represented by "N" (any nucleotide. This residue is missing in the coordinates and the chemical is an ...Details: The actual RNA sequence is: ACAUGAGGAUCACCCAUGUAAUCUCUUUCAAAAAA(2PU)ACAAAAAAAA. (2PU) is represented by "N" (any nucleotide. This residue is missing in the coordinates and the chemical is an internal 2' pyrene modified uridine Number of copies: 2 |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 14.605818 KDa |
| Sequence | String: ACAUGAGGAU CACCCAUGUA AUCUCUUUCA AAAAA(N)ACAA AAAAAA |
-Macromolecule #5: ZINC ION
| Macromolecule | Name: ZINC ION / type: ligand / ID: 5 / Number of copies: 2 / Formula: ZN |
|---|---|
| Molecular weight | Theoretical: 65.409 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 8 mg/mL |
|---|---|
| Buffer | pH: 8 Details: 20 mM Tris-Cl pH 8.0, 50 mM NaCl, 0.1 mM TCEP supplemented with 0.02% (v/v) IGEPAL CA-630 |
| Grid | Model: UltrAuFoil R1.2/1.3 / Material: GOLD / Mesh: 300 / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 295 K / Instrument: FEI VITROBOT MARK IV / Details: 30 s wait time, blot for 2.5 s before plunging. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Average electron dose: 67.6 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: OTHER / Details: Ab initio model from cryosparc |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 4.06 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: RELION Details: A total of 618,412 particles were used for the consensus reconstruction with an overall resolution of 4.06 Angstrom (FSC 0.143 cut off). Focused 3D classification and local refinement were ...Details: A total of 618,412 particles were used for the consensus reconstruction with an overall resolution of 4.06 Angstrom (FSC 0.143 cut off). Focused 3D classification and local refinement were performed in several regions of the complex. A composite map was generated using focused reconstructions of ZCCHC8 HD/KID-MTR4 KOW (3.26 Angstrom, FSC = 0.143; 117,561 particles), protomer A MTR4 (3.42 Angstrom, FSC = 0.143; 225,213 particles), protomer B MTR4 (3.54 Angstrom, FSC = 0.143; 236,602 particles), protomer A MTR4 core-ZCCHC8 PSP-RBM7 RRM (4.06 Angstrom, FSC = 0.143; 44,800 particles), and protomer B MTR4 core-ZCCHC8 PSP-RBM7 RRM (4.4 Angstrom, FSC = 0.143 37,088 particles). Number images used: 618412 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller











 X (Sec.)
X (Sec.) Y (Row.)
Y (Row.) Z (Col.)
Z (Col.)