+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-24840 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Human KATP channel in open conformation, focused on SUR | |||||||||
 Map data Map data | focused on KATP SUR after sharpening (B-factor 82.8) | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | ion channel / KATP / ATP-sensitive potassium channel / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationnegative regulation of neuroblast migration / positive regulation of uterine smooth muscle relaxation / ATP sensitive Potassium channels / Defective ABCC8 can cause hypo- and hyper-glycemias / potassium ion-transporting ATPase complex / negative regulation of blood-brain barrier permeability / ATP-activated inward rectifier potassium channel activity / glutamate secretion, neurotransmission / inward rectifying potassium channel / sulfonylurea receptor activity ...negative regulation of neuroblast migration / positive regulation of uterine smooth muscle relaxation / ATP sensitive Potassium channels / Defective ABCC8 can cause hypo- and hyper-glycemias / potassium ion-transporting ATPase complex / negative regulation of blood-brain barrier permeability / ATP-activated inward rectifier potassium channel activity / glutamate secretion, neurotransmission / inward rectifying potassium channel / sulfonylurea receptor activity / positive regulation of tight junction disassembly / positive regulation of potassium ion transport / ATPase-coupled monoatomic cation transmembrane transporter activity / response to pH / negative regulation of low-density lipoprotein particle clearance / : / negative regulation of glial cell proliferation / response to zinc ion / neuromuscular process / intracellular glucose homeostasis / potassium ion import across plasma membrane / action potential / ATPase-coupled transmembrane transporter activity / potassium channel activity / positive regulation of insulin secretion involved in cellular response to glucose stimulus / ABC-type transporter activity / cellular response to nutrient levels / potassium ion transmembrane transport / negative regulation of angiogenesis / Regulation of insulin secretion / female pregnancy / negative regulation of insulin secretion / response to insulin / ADP binding / sarcolemma / visual learning / potassium ion transport / transmembrane transport / memory / positive regulation of tumor necrosis factor production / synaptic vesicle membrane / presynaptic membrane / response to lipopolysaccharide / transmembrane transporter binding / response to xenobiotic stimulus / ATP hydrolysis activity / ATP binding / metal ion binding / membrane / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.3 Å | |||||||||
 Authors Authors | Zhao C / MacKinnon R | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2021 Journal: Proc Natl Acad Sci U S A / Year: 2021Title: Molecular structure of an open human K channel. Authors: Chen Zhao / Roderick MacKinnon /  Abstract: K channels are metabolic sensors that translate intracellular ATP/ADP balance into membrane excitability. The molecular composition of K includes an inward-rectifier potassium channel (Kir) and an ...K channels are metabolic sensors that translate intracellular ATP/ADP balance into membrane excitability. The molecular composition of K includes an inward-rectifier potassium channel (Kir) and an ABC transporter-like sulfonylurea receptor (SUR). Although structures of K have been determined in many conformations, in all cases, the pore in Kir is closed. Here, we describe human pancreatic K (hK) structures with an open pore at 3.1- to 4.0-Å resolution using single-particle cryo-electron microscopy (cryo-EM). Pore opening is associated with coordinated structural changes within the ATP-binding site and the channel gate in Kir. Conformational changes in SUR are also observed, resulting in an area reduction of contact surfaces between SUR and Kir. We also observe that pancreatic hK exhibits the unique (among inward-rectifier channels) property of PIP-independent opening, which appears to be correlated with a docked cytoplasmic domain in the absence of PIP. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_24840.map.gz emd_24840.map.gz | 173.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-24840-v30.xml emd-24840-v30.xml emd-24840.xml emd-24840.xml | 14.8 KB 14.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_24840_fsc.xml emd_24840_fsc.xml | 12.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_24840.png emd_24840.png | 150.3 KB | ||
| Filedesc metadata |  emd-24840.cif.gz emd-24840.cif.gz | 6.8 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-24840 http://ftp.pdbj.org/pub/emdb/structures/EMD-24840 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-24840 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-24840 | HTTPS FTP |
-Validation report
| Summary document |  emd_24840_validation.pdf.gz emd_24840_validation.pdf.gz | 576.5 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_24840_full_validation.pdf.gz emd_24840_full_validation.pdf.gz | 576.1 KB | Display | |
| Data in XML |  emd_24840_validation.xml.gz emd_24840_validation.xml.gz | 13.2 KB | Display | |
| Data in CIF |  emd_24840_validation.cif.gz emd_24840_validation.cif.gz | 18 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-24840 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-24840 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-24840 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-24840 | HTTPS FTP |
-Related structure data
| Related structure data |  7s5vMC  7s5tC  7s5xC  7s5yC  7s5zC  7s60C  7s61C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_24840.map.gz / Format: CCP4 / Size: 184 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_24840.map.gz / Format: CCP4 / Size: 184 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | focused on KATP SUR after sharpening (B-factor 82.8) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.3 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
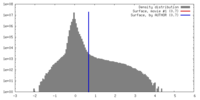
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Human KATP channel composed of Kir6.2 (C166S G334D) and SUR1
| Entire | Name: Human KATP channel composed of Kir6.2 (C166S G334D) and SUR1 |
|---|---|
| Components |
|
-Supramolecule #1: Human KATP channel composed of Kir6.2 (C166S G334D) and SUR1
| Supramolecule | Name: Human KATP channel composed of Kir6.2 (C166S G334D) and SUR1 type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 881.87 kDa/nm |
-Macromolecule #1: ATP-binding cassette sub-family C member 8
| Macromolecule | Name: ATP-binding cassette sub-family C member 8 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 177.106078 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: GPLAFCGSEN HSAAYRVDQG VLNNGCFVDA LNVVPHVFLL FITFPILFIG WGSQSSKVHI HHSTWLHFPG HNLRWILTFM LLFVLVCEI AEGILSDGVT ESHHLHLYMP AGMAFMAAVT SVVYYHNIET SNFPKLLIAL LVYWTLAFIT KTIKFVKFLD H AIGFSQLR ...String: GPLAFCGSEN HSAAYRVDQG VLNNGCFVDA LNVVPHVFLL FITFPILFIG WGSQSSKVHI HHSTWLHFPG HNLRWILTFM LLFVLVCEI AEGILSDGVT ESHHLHLYMP AGMAFMAAVT SVVYYHNIET SNFPKLLIAL LVYWTLAFIT KTIKFVKFLD H AIGFSQLR FCLTGLLVIL YGMLLLVEVN VIRVRRYIFF KTPREVKPPE DLQDLGVRFL QPFVNLLSKG TYWWMNAFIK TA HKKPIDL RAIGKLPIAM RALTNYQRLC EAFDAQVRKD IQGTQGARAI WQALSHAFGR RLVLSSTFRI LADLLGFAGP LCI FGIVDH LGKENDVFQP KTQFLGVYFV SSQEFLANAY VLAVLLFLAL LLQRTFLQAS YYVAIETGIN LRGAIQTKIY NKIM HLSTS NLSMGEMTAG QICNLVAIDT NQLMWFFFLC PNLWAMPVQI IVGVILLYYI LGVSALIGAA VIILLAPVQY FVATK LSQA QRSTLEYSNE RLKQTNEMLR GIKLLKLYAW ENIFRTRVET TRRKEMTSLR AFAIYTSISI FMNTAIPIAA VLITFV GHV SFFKEADFSP SVAFASLSLF HILVTPLFLL SSVVRSTVKA LVSVQKLSEF LSSAEIREEQ CAPHEPTPQG PASKYQA VP LRVVNRKRPA REDCRGLTGP LQSLVPSADG DADNCCVQIM GGYFTWTPDG IPTLSNITIR IPRGQLTMIV GQVGCGKS S LLLAALGEMQ KVSGAVFWSS LPDSEIGEDP SPERETATDL DIRKRGPVAY ASQKPWLLNA TVEENIIFES PFNKQRYKM VIEACSLQPD IDILPHGDQT QIGERGINLS GGQRQRISVA RALYQHANVV FLDDPFSALD IHLSDHLMQA GILELLRDDK RTVVLVTHK LQYLPHADWI IAMKDGTIQR EGTLKDFQRS ECQLFEHWKT LMNRQDQELE KETVTERKAT EPPQGLSRAM S SRDGLLQD EEEEEEEAAE SEEDDNLSSM LHQRAEIPWR ACAKYLSSAG ILLLSLLVFS QLLKHMVLVA IDYWLAKWTD SA LTLTPAA RNCSLSQECT LDQTVYAMVF TVLCSLGIVL CLVTSVTVEW TGLKVAKRLH RSLLNRIILA PMRFFETTPL GSI LNRFSS DCNTIDQHIP STLECLSRST LLCVSALAVI SYVTPVFLVA LLPLAIVCYF IQKYFRVASR DLQQLDDTTQ LPLL SHFAE TVEGLTTIRA FRYEARFQQK LLEYTDSNNI ASLFLTAANR WLEVRMEYIG ACVVLIAAVT SISNSLHREL SAGLV GLGL TYALMVSNYL NWMVRNLADM ELQLGAVKRI HGLLKTEAES YEGLLAPSLI PKNWPDQGKI QIQNLSVRYD SSLKPV LKH VNALIAPGQK IGICGRTGSG KSSFSLAFFR MVDTFEGHII IDGIDIAKLP LHTLRSRLSI ILQDPVLFSG TIRFNLD PE RKCSDSTLWE ALEIAQLKLV VKALPGGLDA IITEGGENFS QGQRQLFCLA RAFVRKTSIF IMDEATASID MATENILQ K VVMTAFADRT VVTIAHRVHT ILSADLVIVL KRGAILEFDK PEKLLSRKDS VFASFVRADK UniProtKB: ATP-binding cassette sub-family C member 8 |
-Macromolecule #2: ADENOSINE-5'-DIPHOSPHATE
| Macromolecule | Name: ADENOSINE-5'-DIPHOSPHATE / type: ligand / ID: 2 / Number of copies: 1 / Formula: ADP |
|---|---|
| Molecular weight | Theoretical: 427.201 Da |
| Chemical component information |  ChemComp-ADP: |
-Macromolecule #3: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 3 / Number of copies: 2 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Macromolecule #4: ADENOSINE-5'-TRIPHOSPHATE
| Macromolecule | Name: ADENOSINE-5'-TRIPHOSPHATE / type: ligand / ID: 4 / Number of copies: 1 / Formula: ATP |
|---|---|
| Molecular weight | Theoretical: 507.181 Da |
| Chemical component information |  ChemComp-ATP: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 6.83 mg/mL |
|---|---|
| Buffer | pH: 8.5 |
| Grid | Model: Quantifoil R1.2/1.3 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 289 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Average electron dose: 57.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller



























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)