[English] 日本語
 Yorodumi
Yorodumi- EMDB-24620: Negative stain electron microscopy single particle reconstruction... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-24620 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Negative stain electron microscopy single particle reconstruction of monoclonal antibody DH1025.1 Fab in complex with HIV-1 Env CH505 SOSIP trimer | |||||||||
 Map data Map data | Final sharpened map of negative stain electron microscopy single particle reconstruction of monoclonal antibody DH1025.1 Fab in complex with HIV-1 Env CH505 SOSIP trimer. | |||||||||
 Sample Sample |
| |||||||||
| Function / homology |  Function and homology information Function and homology informationmembrane fusion involved in viral entry into host cell / symbiont-mediated perturbation of host defense response / positive regulation of plasma membrane raft polarization / positive regulation of receptor clustering / host cell endosome membrane / clathrin-dependent endocytosis of virus by host cell / viral protein processing / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / viral envelope ...membrane fusion involved in viral entry into host cell / symbiont-mediated perturbation of host defense response / positive regulation of plasma membrane raft polarization / positive regulation of receptor clustering / host cell endosome membrane / clathrin-dependent endocytosis of virus by host cell / viral protein processing / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / viral envelope / symbiont entry into host cell / virion attachment to host cell / host cell plasma membrane / virion membrane / structural molecule activity / membrane Similarity search - Function | |||||||||
| Biological species |  Human immunodeficiency virus 2 / Human immunodeficiency virus 2 /    Human immunodeficiency virus 1 Human immunodeficiency virus 1 | |||||||||
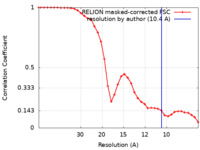
| Method | single particle reconstruction / negative staining / Resolution: 10.4 Å | |||||||||
 Authors Authors | Edwards RJ / Mansouri K | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Sci Transl Med / Year: 2022 Journal: Sci Transl Med / Year: 2022Title: Stabilized HIV-1 envelope immunization induces neutralizing antibodies to the CD4bs and protects macaques against mucosal infection. Authors: Kevin O Saunders / Robert J Edwards / Kedamawit Tilahun / Kartik Manne / Xiaozhi Lu / Derek W Cain / Kevin Wiehe / Wilton B Williams / Katayoun Mansouri / Giovanna E Hernandez / Laura ...Authors: Kevin O Saunders / Robert J Edwards / Kedamawit Tilahun / Kartik Manne / Xiaozhi Lu / Derek W Cain / Kevin Wiehe / Wilton B Williams / Katayoun Mansouri / Giovanna E Hernandez / Laura Sutherland / Richard Scearce / Robert Parks / Maggie Barr / Todd DeMarco / Chloe M Eater / Amanda Eaton / Georgeanna Morton / Benjamin Mildenberg / Yunfei Wang / R Wes Rountree / Mark A Tomai / Christopher B Fox / M Anthony Moody / S Munir Alam / Sampa Santra / Mark G Lewis / Thomas N Denny / George M Shaw / David C Montefiori / Priyamvada Acharya / Barton F Haynes /  Abstract: A successful HIV-1 vaccine will require induction of a polyclonal neutralizing antibody (nAb) response, yet vaccine-mediated induction of such a response in primates remains a challenge. We found ...A successful HIV-1 vaccine will require induction of a polyclonal neutralizing antibody (nAb) response, yet vaccine-mediated induction of such a response in primates remains a challenge. We found that a stabilized HIV-1 CH505 envelope (Env) trimer formulated with a Toll-like receptor 7/8 agonist induced potent HIV-1 polyclonal nAbs that correlated with protection from homologous simian-human immunodeficiency virus (SHIV) infection. The serum dilution that neutralized 50% of virus replication (ID titer) required to protect 90% of macaques was 1:364 against the challenge virus grown in primary rhesus CD4 T cells. Structural analyses of vaccine-induced nAbs demonstrated targeting of the Env CD4 binding site or the N156 glycan and the third variable loop base. Autologous nAb specificities similar to those elicited in macaques by vaccination were isolated from the human living with HIV from which the CH505 Env immunogen was derived. CH505 viral isolates were isolated that mutated the V1 to escape both the infection-induced and vaccine-induced antibodies. These results define the specificities of a vaccine-induced nAb response and the protective titers of HIV-1 vaccine-induced nAbs required to protect nonhuman primates from low-dose mucosal challenge by SHIVs bearing a primary transmitted/founder Env. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_24620.map.gz emd_24620.map.gz | 3.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-24620-v30.xml emd-24620-v30.xml emd-24620.xml emd-24620.xml | 31.7 KB 31.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_24620_fsc.xml emd_24620_fsc.xml | 3.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_24620.png emd_24620.png | 77.2 KB | ||
| Others |  emd_24620_additional_1.map.gz emd_24620_additional_1.map.gz emd_24620_half_map_1.map.gz emd_24620_half_map_1.map.gz emd_24620_half_map_2.map.gz emd_24620_half_map_2.map.gz | 2.4 MB 2.4 MB 2.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-24620 http://ftp.pdbj.org/pub/emdb/structures/EMD-24620 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-24620 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-24620 | HTTPS FTP |
-Related structure data
| Related structure data | C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_24620.map.gz / Format: CCP4 / Size: 3.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_24620.map.gz / Format: CCP4 / Size: 3.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
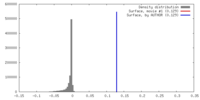
| Annotation | Final sharpened map of negative stain electron microscopy single particle reconstruction of monoclonal antibody DH1025.1 Fab in complex with HIV-1 Env CH505 SOSIP trimer. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 4.02 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: Raw, unsharpened map of negative stain electron microscopy...
| File | emd_24620_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
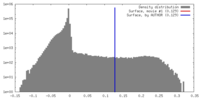
| Annotation | Raw, unsharpened map of negative stain electron microscopy single particle reconstruction of monoclonal antibody DH1025.1 Fab in complex with HIV-1 Env CH505 SOSIP trimer. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half-map 1 of negative stain electron microscopy single...
| File | emd_24620_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
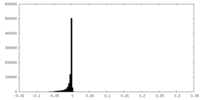
| Annotation | Half-map 1 of negative stain electron microscopy single particle reconstruction of monoclonal antibody DH1025.1 Fab in complex with HIV-1 Env CH505 SOSIP trimer. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half-map 2 of negative stain electron microscopy single...
| File | emd_24620_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
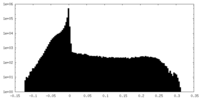
| Annotation | Half-map 2 of negative stain electron microscopy single particle reconstruction of monoclonal antibody DH1025.1 Fab in complex with HIV-1 Env CH505 SOSIP trimer. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Complex of HIV Env CH505TFchim.6R.SOSIP.664.v4.1 with DH1025.1 an...
| Entire | Name: Complex of HIV Env CH505TFchim.6R.SOSIP.664.v4.1 with DH1025.1 antibody Fab |
|---|---|
| Components |
|
-Supramolecule #1: Complex of HIV Env CH505TFchim.6R.SOSIP.664.v4.1 with DH1025.1 an...
| Supramolecule | Name: Complex of HIV Env CH505TFchim.6R.SOSIP.664.v4.1 with DH1025.1 antibody Fab type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Human immunodeficiency virus 2 Human immunodeficiency virus 2 |
| Molecular weight | Theoretical: 23 KDa |
-Supramolecule #2: HIV Env CH505TFchim.6R.SOSIP.664.v4.1 trimer
| Supramolecule | Name: HIV Env CH505TFchim.6R.SOSIP.664.v4.1 trimer / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Human immunodeficiency virus 2 Human immunodeficiency virus 2 |
-Supramolecule #3: DH1025.1 Antibody Fab
| Supramolecule | Name: DH1025.1 Antibody Fab / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #2-#3 |
|---|---|
| Source (natural) | Organism:  |
-Supramolecule #4: DH1025.1 Antibody Fab Heavy Chain
| Supramolecule | Name: DH1025.1 Antibody Fab Heavy Chain / type: complex / ID: 4 / Parent: 3 / Macromolecule list: #2 |
|---|---|
| Source (natural) | Organism:  |
-Supramolecule #5: DH1025.1 Antibody Fab Light Chain
| Supramolecule | Name: DH1025.1 Antibody Fab Light Chain / type: complex / ID: 5 / Parent: 3 / Macromolecule list: #3 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: HIV Env CH505TFchim.6R.SOSIP.664.v4.1
| Macromolecule | Name: HIV Env CH505TFchim.6R.SOSIP.664.v4.1 / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Human immunodeficiency virus 1 Human immunodeficiency virus 1 |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: AENLWVTVYY GVPVWKEAKT TLFCASDAKA YEKEVHNVWA THACVPTDPN PQEMVLKNVT ENFNMWKNDM VDQMHEDVIS LWDQSLKPCV KLTPLCVTLN CTNATASNSS IIEGMKNCSF NITTELRDKR EKKNALFYKL DIVQLDGNSS QYRLINCNTS VITQACPKVS ...String: AENLWVTVYY GVPVWKEAKT TLFCASDAKA YEKEVHNVWA THACVPTDPN PQEMVLKNVT ENFNMWKNDM VDQMHEDVIS LWDQSLKPCV KLTPLCVTLN CTNATASNSS IIEGMKNCSF NITTELRDKR EKKNALFYKL DIVQLDGNSS QYRLINCNTS VITQACPKVS FDPIPIHYCA PAGYAILKCN NKTFTGTGPC NNVSTVQCTH GIKPVVSTQL LLNGSLAEGE IIIRSENITN NVKTIIVHLN ESVKIECTRP NNKTRTSIRI GPGQAFYATG QVIGDIREAY CNINESKWNE TLQRVSKKLK EYFPHKNITF QPSSGGDLEI TTHSFNCGGE FFYCNTSSLF NRTYMANSTD MANSTETNST RTITIHCRIK QIINMWQEVG RAMYAPPIAG NITCISNITG LLLTRDGGKN NTETFRPGGG NMKDNWRSEL YKYKVVKIEP LGVAPTRCKR RVVGRRRRRR AVGIGAVFLG FLGAAGSTMG AASMTLTVQA RNLLSGIVQQ QSNLLRAPEA QQHLLKLTVW GIKQLQARVL AVERYLRDQQ LLGIWGCSGK LICCTNVPWN SSWSNRNLSE IWDNMTWLQW DKEISNYTQI IYGLLEESQN QQEKNEQDLL ALD |
-Macromolecule #2: DH1025.1 Antibody Fab Heavy Chain
| Macromolecule | Name: DH1025.1 Antibody Fab Heavy Chain / type: protein_or_peptide / ID: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: QVHLQESGPG LVKPSETLSL TCAVSGGSVS YYNWWSWIRQ PPGKGLEWIG YISGSSGNTY YSPSLKSRVT ISTDTSKNQF SLRLSSVTAA DTAVYYCASV LQYLDWVLPD RVGEVHWFDV WGPGVLVSVS S astkg ps vfplapss r stsestaal gclvkdyfpe ...String: QVHLQESGPG LVKPSETLSL TCAVSGGSVS YYNWWSWIRQ PPGKGLEWIG YISGSSGNTY YSPSLKSRVT ISTDTSKNQF SLRLSSVTAA DTAVYYCASV LQYLDWVLPD RVGEVHWFDV WGPGVLVSVS S astkg ps vfplapss r stsestaal gclvkdyfpe pvtvswnsg s ltsgvhtf pa vlqssgl ysl ssvvtv psss lgtqt yvcnv nhkp sntkvd krv eiktcgg |
-Macromolecule #3: DH1025.1 Antibody Fab Light Chain
| Macromolecule | Name: DH1025.1 Antibody Fab Light Chain / type: protein_or_peptide / ID: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: DIQMTQSPSS LSASVGDRVT ISCQASQGIS SRLAWYQQRP GKAPKLLIHT ASSLQSGVPS RFSGSGSGTE FTLTISSLQP EDFATYYCQQ HHSHPLTFGG GTKVEIK lf ppsseelq a nkatlvcli sdfypgvvkv awkadgsav n agvetttp sk qsnnkya ass ...String: DIQMTQSPSS LSASVGDRVT ISCQASQGIS SRLAWYQQRP GKAPKLLIHT ASSLQSGVPS RFSGSGSGTE FTLTISSLQP EDFATYYCQQ HHSHPLTFGG GTKVEIK lf ppsseelq a nkatlvcli sdfypgvvkv awkadgsav n agvetttp sk qsnnkya ass ylslts dqwk shksy scqvt hegs tvektv apa ecs |
-Experimental details
-Structure determination
| Method | negative staining |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.4 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.4 Component:
| ||||||||||||
| Staining | Type: NEGATIVE / Material: uranyl formate Details: For negative staining, 5 microliter of the diluted filter retentate was applied to a freshly glow-discharged, 300 mesh, carbon-coated EM grid and incubated 10-12 s, then rinsed with 70 ...Details: For negative staining, 5 microliter of the diluted filter retentate was applied to a freshly glow-discharged, 300 mesh, carbon-coated EM grid and incubated 10-12 s, then rinsed with 70 microliter of 2% uranyl formate delivered in one smooth ejection from a pipet. The drop of uranyl formate remaining on the grid was allowed to incubate for 60 s, then the grid was blotted with filter paper and allowed to air dry. | ||||||||||||
| Grid | Model: Homemade / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: CONTINUOUS / Support film - Film thickness: 5.0 nm / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. | ||||||||||||
| Details | Frozen aliquots of SOSIP and Fab were thawed in an Aluminum block at room temperature for 2-3 minutes, then 10 microgram of SOSIP were mixed with 36 microgram Fab (~1:15 molar ratio) and diluted to 100 microliter final volume with HEPES-buffered saline (HBS), containing 20 mM HEPES buffer and 150 mM NaCl, pH 7.4, augmented with 5% glycerol. Mixture was incubated at 22 degrees C overnight. The following day, a 0.08% glutaraldehyde fixative was prepared by diluting an 8% stock 1/100 with HBS. Then 400 microliter of fixative was added to the SOSIP-Fab mixture and incubated 5 min at room temperature to crosslink the complex, followed by addition of 40 microliter of a 1 M Tris solution, pH 7.4 to quench any unreacted glutaraldehyde. The reaction mixture was then transferred to a 0.5-ml Amicon Ultra centrifugal filter device with a 100-kDa cutoff filter (Millipore, #UFC510096) and spun 10 min at 14,000 g in a benchtop centrifuge held at 20 degrees C to remove excess Fab. The filter retentate, usually ~25 microliter, was collected and diluted 1:1 with HBS augmented with 5% glycerol. |
- Electron microscopy
Electron microscopy
| Microscope | FEI/PHILIPS EM420 |
|---|---|
| Image recording | Film or detector model: OTHER / Digitization - Dimensions - Width: 2048 pixel / Digitization - Dimensions - Height: 2048 pixel / Number grids imaged: 1 / Number real images: 102 / Average exposure time: 0.25 sec. / Average electron dose: 32.0 e/Å2 Details: Images were collected with a minimal dose approach using a manually controlled low dose unit to keep the beam blanked except during initial grid orientation and image collection. To create ...Details: Images were collected with a minimal dose approach using a manually controlled low dose unit to keep the beam blanked except during initial grid orientation and image collection. To create an initial atlas, the entire grid was imaged once at 49x magnification at very low beam intensity by decreasing the emission current, strongly focusing C1 (Spot Size 6) and defocusing C2 (Intensity knob fully clockwise). The stage was then quickly translated to center the desired gridsquare, with an estimated exposure of < 2 s at this very low illumination level, at which point the beam was blanked. A 3,300x magnification was then selected so that a single gridsquare approximately fills the scintillator screen. The beam was then unblanked and the stage manually tilted and the eucentric height quickly adjusted by noting the movement of the gridbars, and then the stage was translated to center on the lower-left corner of the gridsquare, with an estimated exposure of < 2 s at this very low illumination level, at which point the beam was blanked. A magnfication of 82,000x was then selected and C2 focused by a known number of clicks of the Intensity knob so that the beam would approximately fill the scintillator screen. The beam was then unblanked, the objective lens focused by eye and then adjusted to approximately 0.5 micron underfocused, and C2 adjusted so the beam just filled the scintillator screen. Because this focusing step involved a long exposure to the focused beam, no data was collected in this area. For data collection, the stage was translated blindly, with the beam blanked, by noting the micrometer markings on the left-hand stage control, to move to an un-irradiated area that was approximately two screen-widths vertically away from the initial position. The beam was then unblanked, an image captured with 0.25 s exposure to the focused beam without any adjustment of the objective lens focus, and the beam then re-blanked. The FFT of the collected image was examined and the position of the first Thon ring noted and the objective lens focus adjusted if needed for subsequent images. The stage was then moved blindly in the same direction to an un-irradiated area and a new image collected. In this way images were collected until the gridbar was encountered, at which point the beam was unblanked and the stage moved so that the gridbar occluded all of the beam except a small sliver of the gridsquare at the bottom or top of the screen, and the right hand stage control was used to translate the specimen to a new location at least two screen widths away horizontally. In this way the entire gridsquare could be imaged in a rastered fashion with each area only irradiated with the full strength beam during the 0.25 s image capture. |
| Electron beam | Acceleration voltage: 120 kV / Electron source: LAB6 |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.0 mm / Nominal defocus max: 1.5 µm / Nominal defocus min: 0.2 µm / Nominal magnification: 82000 |
| Sample stage | Specimen holder model: SIDE ENTRY, EUCENTRIC / Cooling holder cryogen: NITROGEN |
 Movie
Movie Controller
Controller


 UCSF Chimera
UCSF Chimera















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)