[English] 日本語
 Yorodumi
Yorodumi- EMDB-24319: Structure of the SARS-CoV-2 S 6P trimer in complex with neutraliz... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-24319 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of the SARS-CoV-2 S 6P trimer in complex with neutralizing antibody C051 | |||||||||
 Map data Map data | Sharpened map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | SARS-CoV-2 / receptor binding domain / RBD / neutralizing antibody / COVID-19 / spike / ANTIVIRAL PROTEIN / VIRAL PROTEIN-IMMUNE SYSTEM complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationsymbiont-mediated disruption of host tissue / Maturation of spike protein / Translation of Structural Proteins / Virion Assembly and Release / host cell surface / host extracellular space / symbiont-mediated-mediated suppression of host tetherin activity / Induction of Cell-Cell Fusion / structural constituent of virion / membrane fusion ...symbiont-mediated disruption of host tissue / Maturation of spike protein / Translation of Structural Proteins / Virion Assembly and Release / host cell surface / host extracellular space / symbiont-mediated-mediated suppression of host tetherin activity / Induction of Cell-Cell Fusion / structural constituent of virion / membrane fusion / Attachment and Entry / entry receptor-mediated virion attachment to host cell / host cell endoplasmic reticulum-Golgi intermediate compartment membrane / positive regulation of viral entry into host cell / receptor-mediated virion attachment to host cell / host cell surface receptor binding / symbiont-mediated suppression of host innate immune response / endocytosis involved in viral entry into host cell / receptor ligand activity / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / viral envelope / symbiont entry into host cell / virion attachment to host cell / host cell plasma membrane / SARS-CoV-2 activates/modulates innate and adaptive immune responses / virion membrane / identical protein binding / membrane / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /  | |||||||||
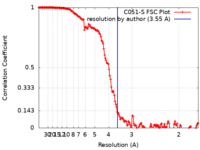
| Method | single particle reconstruction / cryo EM / Resolution: 3.55 Å | |||||||||
 Authors Authors | Barnes CO / Bjorkman PJ | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation | Journal: bioRxiv / Year: 2021 Title: Development of potency, breadth and resilience to viral escape mutations in SARS-CoV-2 neutralizing antibodies. Authors: Frauke Muecksch / Yiska Weisblum / Christopher O Barnes / Fabian Schmidt / Dennis Schaefer-Babajew / Julio C C Lorenzi / Andrew I Flyak / Andrew T DeLaitsch / Kathryn E Huey-Tubman / Shurong ...Authors: Frauke Muecksch / Yiska Weisblum / Christopher O Barnes / Fabian Schmidt / Dennis Schaefer-Babajew / Julio C C Lorenzi / Andrew I Flyak / Andrew T DeLaitsch / Kathryn E Huey-Tubman / Shurong Hou / Celia A Schiffer / Christian Gaebler / Zijun Wang / Justin Da Silva / Daniel Poston / Shlomo Finkin / Alice Cho / Melissa Cipolla / Thiago Y Oliveira / Katrina G Millard / Victor Ramos / Anna Gazumyan / Magdalena Rutkowska / Marina Caskey / Michel C Nussenzweig / Pamela J Bjorkman / Theodora Hatziioannou / Paul D Bieniasz /  Abstract: Antibodies elicited in response to infection undergo somatic mutation in germinal centers that can result in higher affinity for the cognate antigen. To determine the effects of somatic mutation on ...Antibodies elicited in response to infection undergo somatic mutation in germinal centers that can result in higher affinity for the cognate antigen. To determine the effects of somatic mutation on the properties of SARS-CoV-2 spike receptor-binding domain (RBD)-specific antibodies, we analyzed six independent antibody lineages. As well as increased neutralization potency, antibody evolution changed pathways for acquisition of resistance and, in some cases, restricted the range of neutralization escape options. For some antibodies, maturation apparently imposed a requirement for multiple spike mutations to enable escape. For certain antibody lineages, maturation enabled neutralization of circulating SARS-CoV-2 variants of concern and heterologous sarbecoviruses. Antibody-antigen structures revealed that these properties resulted from substitutions that allowed additional variability at the interface with the RBD. These findings suggest that increasing antibody diversity through prolonged or repeated antigen exposure may improve protection against diversifying SARS-CoV-2 populations, and perhaps against other pandemic threat coronaviruses. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_24319.map.gz emd_24319.map.gz | 290.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-24319-v30.xml emd-24319-v30.xml emd-24319.xml emd-24319.xml | 22.4 KB 22.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_24319_fsc.xml emd_24319_fsc.xml | 15.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_24319.png emd_24319.png | 56.3 KB | ||
| Filedesc metadata |  emd-24319.cif.gz emd-24319.cif.gz | 7.7 KB | ||
| Others |  emd_24319_additional_1.map.gz emd_24319_additional_1.map.gz | 151.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-24319 http://ftp.pdbj.org/pub/emdb/structures/EMD-24319 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-24319 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-24319 | HTTPS FTP |
-Related structure data
| Related structure data |  7r8nMC  7n3eC  7n3fC  7n3gC  7n3hC  7n3iC  7r8lC  7r8mC  7r8oC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_24319.map.gz / Format: CCP4 / Size: 307.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_24319.map.gz / Format: CCP4 / Size: 307.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sharpened map | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.869 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: Unsharpened map
| File | emd_24319_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unsharpened map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Ternary complex of SARS-CoV-2 S 6P trimer with C051 Fabs
| Entire | Name: Ternary complex of SARS-CoV-2 S 6P trimer with C051 Fabs |
|---|---|
| Components |
|
-Supramolecule #1: Ternary complex of SARS-CoV-2 S 6P trimer with C051 Fabs
| Supramolecule | Name: Ternary complex of SARS-CoV-2 S 6P trimer with C051 Fabs type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 720 KDa |
-Macromolecule #1: Spike glycoprotein
| Macromolecule | Name: Spike glycoprotein / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 141.157391 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MFVFLVLLPL VSSQCVNLTT RTQLPPAYTN SFTRGVYYPD KVFRSSVLHS TQDLFLPFFS NVTWFHAIHV SGTNGTKRFD NPVLPFNDG VYFASTEKSN IIRGWIFGTT LDSKTQSLLI VNNATNVVIK VCEFQFCNDP FLGVYYHKNN KSWMESEFRV Y SSANNCTF ...String: MFVFLVLLPL VSSQCVNLTT RTQLPPAYTN SFTRGVYYPD KVFRSSVLHS TQDLFLPFFS NVTWFHAIHV SGTNGTKRFD NPVLPFNDG VYFASTEKSN IIRGWIFGTT LDSKTQSLLI VNNATNVVIK VCEFQFCNDP FLGVYYHKNN KSWMESEFRV Y SSANNCTF EYVSQPFLMD LEGKQGNFKN LREFVFKNID GYFKIYSKHT PINLVRDLPQ GFSALEPLVD LPIGINITRF QT LLALHRS YLTPGDSSSG WTAGAAAYYV GYLQPRTFLL KYNENGTITD AVDCALDPLS ETKCTLKSFT VEKGIYQTSN FRV QPTESI VRFPNITNLC PFGEVFNATR FASVYAWNRK RISNCVADYS VLYNSASFST FKCYGVSPTK LNDLCFTNVY ADSF VIRGD EVRQIAPGQT GKIADYNYKL PDDFTGCVIA WNSNNLDSKV GGNYNYLYRL FRKSNLKPFE RDISTEIYQA GSTPC NGVE GFNCYFPLQS YGFQPTNGVG YQPYRVVVLS FELLHAPATV CGPKKSTNLV KNKCVNFNFN GLTGTGVLTE SNKKFL PFQ QFGRDIADTT DAVRDPQTLE ILDITPCSFG GVSVITPGTN TSNQVAVLYQ DVNCTEVPVA IHADQLTPTW RVYSTGS NV FQTRAGCLIG AEHVNNSYEC DIPIGAGICA SYQTQTNSPA SVASQSIIAY TMSLGAENSV AYSNNSIAIP TNFTISVT T EILPVSMTKT SVDCTMYICG DSTECSNLLL QYGSFCTQLN RALTGIAVEQ DKNTQEVFAQ VKQIYKTPPI KDFGGFNFS QILPDPSKPS KRSPIEDLLF NKVTLADAGF IKQYGDCLGD IAARDLICAQ KFNGLTVLPP LLTDEMIAQY TSALLAGTIT SGWTFGAGP ALQIPFPMQM AYRFNGIGVT QNVLYENQKL IANQFNSAIG KIQDSLSSTP SALGKLQDVV NQNAQALNTL V KQLSSNFG AISSVLNDIL SRLDPPEAEV QIDRLITGRL QSLQTYVTQQ LIRAAEIRAS ANLAATKMSE CVLGQSKRVD FC GKGYHLM SFPQSAPHGV VFLHVTYVPA QEKNFTTAPA ICHDGKAHFP REGVFVSNGT HWFVTQRNFY EPQIITTDNT FVS GNCDVV IGIVNNTVYD PLQPELDSFK EELDKYFKNH TSPDVDLGDI SGINASVVNI QKEIDRLNEV AKNLNESLID LQEL GKYEQ YIKWPSGRLV PRGSPGSGYI PEAPRDGQAY VRKDGEWVLL STFLGHHHHH HGLNDIFEAQ KIEWHE UniProtKB: Spike glycoprotein |
-Macromolecule #2: C051 Fab Heavy Chain
| Macromolecule | Name: C051 Fab Heavy Chain / type: protein_or_peptide / ID: 2 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 25.418262 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: EVQLVESGGG LIQAGGSLRL SCAASGFGVR NNYMSWVRQA PGKGLEWVSV IYSGGTTYYA DSVKGRFTIS RDNSKNTVFL QMNSLRAED TAVYYCAREG DVEGFSDLWS GYSRDRYYFD YWGQGTLVTV SSASTKGPSV FPLAPSSKST SGGTAALGCL V KDYFPEPV ...String: EVQLVESGGG LIQAGGSLRL SCAASGFGVR NNYMSWVRQA PGKGLEWVSV IYSGGTTYYA DSVKGRFTIS RDNSKNTVFL QMNSLRAED TAVYYCAREG DVEGFSDLWS GYSRDRYYFD YWGQGTLVTV SSASTKGPSV FPLAPSSKST SGGTAALGCL V KDYFPEPV TVSWNSGALT SGVHTFPAVL QSSGLYSLSS VVTVPSSSLG TQTYICNVNH KPSNTKVDKR VEPKSCDKT |
-Macromolecule #3: C051 Fab Light Chain
| Macromolecule | Name: C051 Fab Light Chain / type: protein_or_peptide / ID: 3 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 22.866094 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: QSVLTQPASV SGSPGQSITF SCTGTSSDVG GYNYVSWYQQ YPGKAPKLLI YDVTNRPSGV SDRFSGSKSG NTASLTISGL QAEDEADYY CSSFTSSNTR VFGTGTKVTV LGQPKAAPSV TLFPPSSEEL QANKATLVCL ISDFYPGAVT VAWKADSSPV K AGVETTTP ...String: QSVLTQPASV SGSPGQSITF SCTGTSSDVG GYNYVSWYQQ YPGKAPKLLI YDVTNRPSGV SDRFSGSKSG NTASLTISGL QAEDEADYY CSSFTSSNTR VFGTGTKVTV LGQPKAAPSV TLFPPSSEEL QANKATLVCL ISDFYPGAVT VAWKADSSPV K AGVETTTP SKQSNNKYAA SSYLSLTPEQ WKSHRSYSCQ VTHEGSTVEK TVAPTECS |
-Macromolecule #5: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 5 / Number of copies: 27 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 3.0 mg/mL | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
| |||||||||
| Grid | Model: UltrAuFoil R1.2/1.3 / Material: GOLD / Mesh: 300 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 60 sec. / Pretreatment - Atmosphere: AIR / Details: .2 mA | |||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 295 K / Instrument: FEI VITROBOT MARK IV / Details: 3s blot, 0 blot force. | |||||||||
| Details | Monodisperse sample |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Number grids imaged: 1 / Number real images: 3402 / Average exposure time: 3.6 sec. / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Calibrated defocus max: 2.0 µm / Calibrated magnification: 45000 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus min: 0.7000000000000001 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller
















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)