+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-24303 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | In situ cryoET map of influenza A hemagglutinin | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
| Biological species |  Influenza A virus (A/Puerto Rico/8/1934(H1N1)) Influenza A virus (A/Puerto Rico/8/1934(H1N1)) | |||||||||
| Method | subtomogram averaging / cryo EM / Resolution: 12.9 Å | |||||||||
 Authors Authors | Huang QY / Song K / Schiffer CA / Somasundaran M | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Structure / Year: 2022 Journal: Structure / Year: 2022Title: Quantitative structural analysis of influenza virus by cryo-electron tomography and convolutional neural networks. Authors: Qiuyu J Huang / Kangkang Song / Chen Xu / Daniel N A Bolon / Jennifer P Wang / Robert W Finberg / Celia A Schiffer / Mohan Somasundaran /  Abstract: Influenza viruses pose severe public health threats globally. Influenza viruses are extensively pleomorphic, in shape, size, and organization of viral proteins. Analysis of influenza morphology and ...Influenza viruses pose severe public health threats globally. Influenza viruses are extensively pleomorphic, in shape, size, and organization of viral proteins. Analysis of influenza morphology and ultrastructure can help elucidate viral structure-function relationships and aid in therapeutics and vaccine development. While cryo-electron tomography (cryoET) can depict the 3D organization of pleomorphic influenza, the low signal-to-noise ratio inherent to cryoET and viral heterogeneity have precluded detailed characterization of influenza viruses. In this report, we leveraged convolutional neural networks and cryoET to characterize the morphological architecture of the A/Puerto Rico/8/34 (H1N1) influenza strain. Our pipeline improved the throughput of cryoET analysis and accurately identified viral components within tomograms. Using this approach, we successfully characterized influenza morphology, glycoprotein density, and conducted subtomogram averaging of influenza glycoproteins. Application of this processing pipeline can aid in the structural characterization of not only influenza viruses, but other pleomorphic viruses and infected cells. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_24303.map.gz emd_24303.map.gz | 24.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-24303-v30.xml emd-24303-v30.xml emd-24303.xml emd-24303.xml | 15.2 KB 15.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_24303_fsc.xml emd_24303_fsc.xml | 9 KB | Display |  FSC data file FSC data file |
| Images |  emd_24303.png emd_24303.png | 25.2 KB | ||
| Others |  emd_24303_half_map_1.map.gz emd_24303_half_map_1.map.gz emd_24303_half_map_2.map.gz emd_24303_half_map_2.map.gz | 11.4 MB 11.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-24303 http://ftp.pdbj.org/pub/emdb/structures/EMD-24303 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-24303 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-24303 | HTTPS FTP |
-Validation report
| Summary document |  emd_24303_validation.pdf.gz emd_24303_validation.pdf.gz | 603.3 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_24303_full_validation.pdf.gz emd_24303_full_validation.pdf.gz | 602.8 KB | Display | |
| Data in XML |  emd_24303_validation.xml.gz emd_24303_validation.xml.gz | 13.1 KB | Display | |
| Data in CIF |  emd_24303_validation.cif.gz emd_24303_validation.cif.gz | 16.6 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-24303 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-24303 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-24303 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-24303 | HTTPS FTP |
-Related structure data
| Similar structure data |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_24303.map.gz / Format: CCP4 / Size: 27 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_24303.map.gz / Format: CCP4 / Size: 27 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.087 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
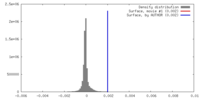
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Half map: #2
| File | emd_24303_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
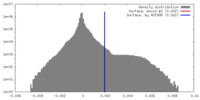
| Density Histograms |
-Half map: #1
| File | emd_24303_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
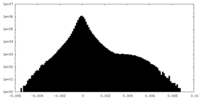
| Density Histograms |
- Sample components
Sample components
-Entire : Influenza A virus (A/Puerto Rico/8/1934(H1N1))
| Entire | Name:  Influenza A virus (A/Puerto Rico/8/1934(H1N1)) Influenza A virus (A/Puerto Rico/8/1934(H1N1)) |
|---|---|
| Components |
|
-Supramolecule #1: Influenza A virus (A/Puerto Rico/8/1934(H1N1))
| Supramolecule | Name: Influenza A virus (A/Puerto Rico/8/1934(H1N1)) / type: virus / ID: 1 / Parent: 0 / Macromolecule list: all / Details: Purchased from Charles River Laboratories. / NCBI-ID: 211044 Sci species name: Influenza A virus (A/Puerto Rico/8/1934(H1N1)) Virus type: VIRION / Virus isolate: STRAIN / Virus enveloped: Yes / Virus empty: No |
|---|---|
| Host (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 198 KDa |
-Macromolecule #1: Influenza A H1 hemagglutinin
| Macromolecule | Name: Influenza A H1 hemagglutinin / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Influenza A virus (A/Puerto Rico/8/1934(H1N1)) Influenza A virus (A/Puerto Rico/8/1934(H1N1)) |
| Sequence | String: MKANLLVLLC ALAAADADTI CIGYHANNST DTVDTVLEKN VTVTHSVNLL EDSHNGKLCR LKGIAPLQLG KCNIAGWLLG NPECDPLLPV RSWSYIVETP NSENGICYPG DFIDYEELRE QLSSVSSFER FEIFPKESSW PNHNTTKGVT AACSHAGKSS FYRNLLWLTE ...String: MKANLLVLLC ALAAADADTI CIGYHANNST DTVDTVLEKN VTVTHSVNLL EDSHNGKLCR LKGIAPLQLG KCNIAGWLLG NPECDPLLPV RSWSYIVETP NSENGICYPG DFIDYEELRE QLSSVSSFER FEIFPKESSW PNHNTTKGVT AACSHAGKSS FYRNLLWLTE KEGSYPKLKN SYVNKKGKEV LVLWGIHHPS NSKDQQNIYQ NENAYVSVVT SNYNRRFTPE IAERPKVRDQ AGRMNYYWTL LKPGDTIIFE ANGNLIAPRY AFALSRGFGS GIITSNASMH ECNTKCQTPL GAINSSLPFQ NIHPVTIGEC PKYVRSAKLR MVTGLRNIPS IQSRGLFGAI AGFIEGGWTG MIDGWYGYHH QNEQGSGYAA DQKSTQNAIN GITNKVNSVI EKMNIQFTAV GKEFNKLEKR MENLNKKVDD GFLDIWTYNA ELLVLLENER TLDFHDSNVK NLYEKVKSQL KNNAKEIGNG CFEFYHKCDN ECMESVRNGT YDYPKYSEES KLNREKVDGV KLESMGIYQI LAIYSTVASS LVLLVSLGAI SFWMCSNGSL QCRICI |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | subtomogram averaging |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1 mg/mL |
|---|---|
| Buffer | pH: 7.4 |
| Grid | Model: Quantifoil R2/2 / Material: COPPER / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE / Instrument: EMS-002 RAPID IMMERSION FREEZER |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 2.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | PDB ID: |
|---|---|
| Refinement | Protocol: FLEXIBLE FIT |
 Movie
Movie Controller
Controller









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)